User:Vand1169/sandbox: Difference between revisions
m Task 11: Disable the categories on this page while it is still a draft, per WP:USERNOCAT, using PC bot (v. 1.7) |
Citation bot (talk | contribs) Add: url-status. Removed parameters. Some additions/deletions were parameter name changes. | Use this bot. Report bugs. | Suggested by AManWithNoPlan | #UCB_CommandLine |
||
| (3 intermediate revisions by one other user not shown) | |||
| Line 8: | Line 8: | ||
==Gene== |
==Gene== |
||
The KIAA0895 gene is located at 7p14.2.[[Image:KIAA0895location.png|thumb|The location of the KIAA0895 gene on chromosome 7|445x445px|alt=]] It can be transcribed into 15 [[alternative splicing|transcript variants]], which in turn can produce |
The KIAA0895 gene is located at 7p14.2.[[Image:KIAA0895location.png|thumb|The location of the KIAA0895 gene on chromosome 7|445x445px|alt=]] It can be transcribed into 15 [[alternative splicing|transcript variants]], which in turn can produce ten different [[isoforms]] of the protein.<ref name="aceview">{{cite web | title = NCBI AceView: KIAA0895| url = https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=human&term=kiaa0895&submit=Go| access-date = 2011-04-24}}</ref> |
||
The genomic DNA is 65,976 base pairs long,<ref name="entrez">{{cite web|url=https://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=retrieve&list_uids=23366|title=Entrez Gene: KIAA0895|access-date=2011-04-24}}</ref> while the longest mRNA that it produces is 4463 bases long. |
The genomic DNA is 65,976 base pairs long,<ref name="entrez">{{cite web|url=https://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=retrieve&list_uids=23366|title=Entrez Gene: KIAA0895|access-date=2011-04-24}}</ref> while the longest mRNA that it produces is 4463 bases long. |
||
| Line 30: | Line 30: | ||
*XP_024302470.1 |
*XP_024302470.1 |
||
*NP_001186637.1 |
*NP_001186637.1 |
||
*NP_001287885.1{{div col end}}A |
*NP_001287885.1{{div col end}}A stem-loop formation for the 5' UTR region shows that there is a lack of conservation, meaning there is some precedence that these stems and loops are not used for translation regulation.<ref name=":1">{{Cite web|url=http://unafold.rna.albany.edu/?q=mfold|title=The Mfold Web Server {{!}} mfold.rit.albany.edu|website=unafold.rna.albany.edu|access-date=2019-04-17}}</ref> The ΔG value was -12.90 kcal/mol with three loops.<ref name=":1" /> |
||
A |
A stem-loop formation for the 3' UTR region shows that there is conservation, meaning there is some precedence that they are used for translation regulation.<ref name=":1" /> The ΔG value was -636.80 kcal/mol.<ref name=":1" /> |
||
==Protein== |
==Protein== |
||
| Line 53: | Line 53: | ||
}} |
}} |
||
===Regions=== |
===Regions=== |
||
<nowiki> </nowiki>LOC23366 contains a protein [[domain of unknown function]] called DUF1704.<ref name="Pfam Sanger">{{cite web|title=Wellcome Trust Sanger Institute, Pfam |url=http://pfam.sanger.ac.uk/ |archive-url=http://webarchive.nationalarchives.gov.uk/20140617135541/http://pfam.sanger.ac.uk/ | |
<nowiki> </nowiki>LOC23366 contains a protein [[domain of unknown function]] called DUF1704.<ref name="Pfam Sanger">{{cite web|title=Wellcome Trust Sanger Institute, Pfam |url=http://pfam.sanger.ac.uk/ |archive-url=http://webarchive.nationalarchives.gov.uk/20140617135541/http://pfam.sanger.ac.uk/ |url-status=dead |archive-date=2014-06-17 |access-date=2011-05-09 }}</ref> |
||
It also contains a region of low complexity from position 120 to position 150 in the protein,<ref name = "Dotlet">{{cite web | title =MyHits Dotlet| url = http://myhits.isb-sib.ch/cgi-bin/dotlet| access-date = 2011-05-09}}</ref> and an [[arginine]]-rich area from position 12 to position 51.<ref name = "Uniprot">{{cite web | title =Uniprot| url = https://www.uniprot.org/uniprot/Q8NCT3| access-date = 2011-05-09}}</ref> |
It also contains a region of low complexity from position 120 to position 150 in the protein,<ref name = "Dotlet">{{cite web | title =MyHits Dotlet| url = http://myhits.isb-sib.ch/cgi-bin/dotlet| access-date = 2011-05-09}}</ref> and an [[arginine]]-rich area from position 12 to position 51.<ref name = "Uniprot">{{cite web | title =Uniprot| url = https://www.uniprot.org/uniprot/Q8NCT3| access-date = 2011-05-09}}</ref> |
||
=== Amino Acid Composition === |
=== Amino Acid Composition === |
||
KIAA0895 is a [[lysine]] and [[arginine]] semi-enriched protein.<ref name=":3">{{Cite web|url=https://www.ebi.ac.uk/Tools/seqstats/saps/|title=SAPS < Sequence Statistics < EMBL-EBI|website=www.ebi.ac.uk|access-date=2019-04-17}}</ref> KIAA0895 is semi-enriched in positively charged lysine and arginine groups, and positively and negatively charged lysine, arginine, [[glutamic acid]] and [[aspartic acid]] groups.<ref name=":3" /> However, KIAA0895 is depleted in non-polar [[alanine]], [[glycine]] and [[proline]] groups.<ref name=":3" /> |
KIAA0895 is a [[lysine]] and [[arginine]] semi-enriched protein.<ref name=":3">{{Cite web|url=https://www.ebi.ac.uk/Tools/seqstats/saps/|title=SAPS < Sequence Statistics < EMBL-EBI|website=www.ebi.ac.uk|access-date=2019-04-17}}</ref> KIAA0895 is semi-enriched in positively charged lysine and arginine groups, and positively and negatively charged lysine, arginine, [[glutamic acid]] and [[aspartic acid]] groups.<ref name=":3" /> However, KIAA0895 is semi-depleted in [[non-polar]] [[alanine]], [[glycine]] and [[proline]] groups.<ref name=":3" /> |
||
The charge distribution analysis shows that there are no negative or mixed charge clusters.<ref name=":3" /> However, there is one positive charge cluster from amino acids 12 to 36.<ref name=":3" /> |
The charge distribution analysis shows that there are no negative or mixed charge clusters.<ref name=":3" /> However, there is one positive charge cluster from amino acids 12 to 36.<ref name=":3" /> |
||
| Line 66: | Line 66: | ||
[[Image:Kiaa0895 phosphorylation sites.gif|thumb|center|400px|The predicted [[serine]], [[threonine]], and [[tyrosine]] phosphorylation sites of the KIAA0895 protein]]There are also predicted to be three [[N-linked glycosylation]] sites.<ref name="NetNGlyc">{{cite web|url=http://www.cbs.dtu.dk/services/NetNGlyc/|title=DTU Center for Biological Sciences, NetNGlyc|access-date=2011-05-09}}</ref> These occur at amino acids 142, 316, and 363. The sequences for these sites are NTS, NVS and NPT, respectfully. N-linked glycosylation has both [[Intrinsic and extrinsic properties|intrinsic and extrinsic]] functions, regulating the migration patterns of cells. |
[[Image:Kiaa0895 phosphorylation sites.gif|thumb|center|400px|The predicted [[serine]], [[threonine]], and [[tyrosine]] phosphorylation sites of the KIAA0895 protein]]There are also predicted to be three [[N-linked glycosylation]] sites.<ref name="NetNGlyc">{{cite web|url=http://www.cbs.dtu.dk/services/NetNGlyc/|title=DTU Center for Biological Sciences, NetNGlyc|access-date=2011-05-09}}</ref> These occur at amino acids 142, 316, and 363. The sequences for these sites are NTS, NVS and NPT, respectfully. N-linked glycosylation has both [[Intrinsic and extrinsic properties|intrinsic and extrinsic]] functions, regulating the migration patterns of cells. |
||
[[File:KIAA0895 N-linked glycosylation.gif|center|thumb|393x393px|The three predicted N-linked glycosylation sites.]] |
[[File:KIAA0895 N-linked glycosylation.gif|center|thumb|393x393px|The three predicted N-linked glycosylation sites.]] |
||
=== Tertiary Structure === |
|||
KIAA0895 has a tertiary structure with [[Alpha helix|alpha helices]] and [[Beta sheet|beta sheets]].<ref>{{Cite web|url=http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index|title=PHYRE2 Protein Fold Recognition Server|website=www.sbg.bio.ic.ac.uk|access-date=2019-04-20}}</ref> |
|||
[[File:KIAA0895TertiaryStructure.png|center|thumb|207x207px|Proposed Tertiary Structure for KIAA0895. Image coloured by rainbow N → C terminus.]] |
|||
<br /> |
|||
=== Interacting Proteins === |
|||
There are three proteins likely to be interacting proteins with KIAA0895. These proteins are [[ELAV-like protein 1|ELAVL1]]<ref>{{Cite web|url=https://mentha.uniroma2.it/|title=mentha: the interactome browser|website=mentha.uniroma2.it|access-date=2019-04-20}}</ref>, vata<ref name=":4">{{Cite web|url=https://www.ebi.ac.uk/intact/|website=www.ebi.ac.uk|access-date=2019-04-20}}</ref>, and glym<ref name=":4" />. These interactions have experimental evidence from the sources provided. |
|||
== Expression == |
== Expression == |
||
KIAA0895 is most commonly found in the [[Testicle|testis]], however it also has a strong expression in the [[kidney]]s, [[adrenal gland]]s, [[brain]], [[ovary]], [[skin]] and [[ |
KIAA0895 is most commonly found in the [[Testicle|testis]], however it also has a strong expression in the [[kidney]]s, [[adrenal gland]]s, [[brain]], [[ovary]], [[skin]], [[stomach]], [[pancreas]], [[bone marrow]], [[Parathyroid gland|parathyroid]], and [[spinal cord]].<ref name=":0">{{Cite web|url=https://www.ncbi.nlm.nih.gov/gene/23366|title=KIAA0895 KIAA0895 [Homo sapiens (human)] - Gene - NCBI|website=www.ncbi.nlm.nih.gov|access-date=2019-02-24}}</ref> [[File:Human tissue-specific expression of KIAA0895.png|thumb|308x308px|The Mean RPKM values for KIAA0895 in 27 different tissue types.|alt=|center]] |
||
==Homologs and Orthologs== |
==Homologs and Orthologs== |
||
KIAA0895 has over 228 [[Homology (biology)|orthologs]].<ref name=":0" /> Orthologs have been found in [[mammals]] and [[eukaryote]]s.<ref name="BLAST">{{cite web|url=http://blast.ncbi.nlm.nih.gov/Blast.cgi|title=NCBI BLAST|access-date=2011-05-09}}</ref> There are [[Homology (biology)|homologs]] in 9 species.<ref name=":0" /> The full list of organisms in which homologs have been found is given below. |
KIAA0895 has over 228 [[Homology (biology)|orthologs]].<ref name=":0" /> Orthologs have been found in [[mammals]] and [[eukaryote]]s.<ref name="BLAST">{{cite web|url=http://blast.ncbi.nlm.nih.gov/Blast.cgi|title=NCBI BLAST|access-date=2011-05-09}}</ref> There are [[Homology (biology)|homologs]] in 9 species.<ref name=":0" /> The full list of organisms in which homologs have been found is given below. |
||
| Line 84: | Line 91: | ||
== Paralogs == |
== Paralogs == |
||
KIAA0895 has 7 paralogs in ''[[Homo sapiens]] |
KIAA0895 has 7 paralogs in ''[[Homo sapiens]]:''<ref name="BLAST" />{{div col|colwidth=20em}} |
||
*''Unnamed protein product'' |
*''Unnamed protein product'' |
||
*''Uncharacterized protein KIAA0895-like'' |
*''Uncharacterized protein KIAA0895-like'' |
||
Latest revision as of 07:46, 8 June 2022
An Error has occurred retrieving Wikidata item for infobox
KIAA0895 is a gene found in Homo sapiens. The gene encodes a protein commonly known as the KIAA0895 protein. It's aliases include hypothetical protein LOC23366, OTTHUMP00000206979, OTTHUMP00000206980, 9530077C05Rik, 1110003N12Rik, Kiaa0895, and mKIAA0895.[1] Research into the KIAA proteins has shown that they are similar to known genes with functions related to cell signaling/communication, cell structure/motility and nucleic acid management.[2]
Gene
[edit]The KIAA0895 gene is located at 7p14.2.
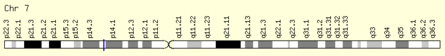
It can be transcribed into 15 transcript variants, which in turn can produce ten different isoforms of the protein.[3]
The genomic DNA is 65,976 base pairs long,[4] while the longest mRNA that it produces is 4463 bases long.
Gene Neighborhood
[edit]KIAA0895 is surrounded by the following genes on chromosome 7:[4]
Transcript
[edit]There are ten different isoforms for KIAA0895.[5]
- NP_001093895.1
- EAW94064.1
- NP_056129.2
- NP_001186636.1
- NP_001186635.1
- XP_005249746.1
- EAW94065.1
- XP_024302470.1
- NP_001186637.1
- NP_001287885.1
A stem-loop formation for the 5' UTR region shows that there is a lack of conservation, meaning there is some precedence that these stems and loops are not used for translation regulation.[6] The ΔG value was -12.90 kcal/mol with three loops.[6]
A stem-loop formation for the 3' UTR region shows that there is conservation, meaning there is some precedence that they are used for translation regulation.[6] The ΔG value was -636.80 kcal/mol.[6]
Protein
[edit]The longest protein isoform that is produced by the KIAA0895 gene is termed LOC23366 isoform 1 and is 520 amino acids long.[7] The predicted molecular weight is 61kDa.[8] Additionally, the theoretical isoelectric point is 10.[8]
| Domain of unknown function 1704 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | DUF1704 | ||||||||
| Pfam | PF08014 | ||||||||
| InterPro | IPR012548 | ||||||||
| |||||||||
Regions
[edit]LOC23366 contains a protein domain of unknown function called DUF1704.[9] It also contains a region of low complexity from position 120 to position 150 in the protein,[10] and an arginine-rich area from position 12 to position 51.[11]
Amino Acid Composition
[edit]KIAA0895 is a lysine and arginine semi-enriched protein.[12] KIAA0895 is semi-enriched in positively charged lysine and arginine groups, and positively and negatively charged lysine, arginine, glutamic acid and aspartic acid groups.[12] However, KIAA0895 is semi-depleted in non-polar alanine, glycine and proline groups.[12]
The charge distribution analysis shows that there are no negative or mixed charge clusters.[12] However, there is one positive charge cluster from amino acids 12 to 36.[12]
Post-translational Modification
[edit]KIAA0895 is predicted to undergo phosphorylation at several serines, threonines, and tyrosines throughout its structure.[13] Phosphorylation at these sites is a form of gene regulation. Phosphorylation results in a conformational change in the structure of many enzymes and receptors. This causes them to become activated or deactivated.

There are also predicted to be three N-linked glycosylation sites.[14] These occur at amino acids 142, 316, and 363. The sequences for these sites are NTS, NVS and NPT, respectfully. N-linked glycosylation has both intrinsic and extrinsic functions, regulating the migration patterns of cells.

Tertiary Structure
[edit]KIAA0895 has a tertiary structure with alpha helices and beta sheets.[15]

Interacting Proteins
[edit]There are three proteins likely to be interacting proteins with KIAA0895. These proteins are ELAVL1[16], vata[17], and glym[17]. These interactions have experimental evidence from the sources provided.
Expression
[edit]KIAA0895 is most commonly found in the testis, however it also has a strong expression in the kidneys, adrenal glands, brain, ovary, skin, stomach, pancreas, bone marrow, parathyroid, and spinal cord.[5]

Homologs and Orthologs
[edit]KIAA0895 has over 228 orthologs.[5] Orthologs have been found in mammals and eukaryotes.[18] There are homologs in 9 species.[5] The full list of organisms in which homologs have been found is given below.
Paralogs
[edit]KIAA0895 has 7 paralogs in Homo sapiens:[18]
- Unnamed protein product
- Uncharacterized protein KIAA0895-like
- hCG28832, isoform CRA_a
- Hypothetical protein
- Unnamed protein product
- hCG38687, isoform CRA_a, partial
- hCG38687, isoform CRA_b
References
[edit]- ^ "GeneCards: KIAA0895 Gene". Retrieved 2011-04-24.
- ^ Nagase T, Ishikawa K, Suyama M, Kikuno R, Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O (December 1998). "Prediction of the coding sequences of unidentified human genes. XII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro". DNA Research. 5 (6): 355–64. doi:10.1093/dnares/5.6.355. PMID 10048485.
- ^ "NCBI AceView: KIAA0895". Retrieved 2011-04-24.
- ^ a b "Entrez Gene: KIAA0895". Retrieved 2011-04-24.
- ^ a b c d "KIAA0895 KIAA0895 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2019-02-24.
- ^ a b c d "The Mfold Web Server | mfold.rit.albany.edu". unafold.rna.albany.edu. Retrieved 2019-04-17.
- ^ "NCBI Protein: LOC23366 isoform 1". Retrieved 2011-05-09.
- ^ a b "ExPASy - Compute pI/Mw tool". web.expasy.org. Retrieved 2019-04-17.
- ^ "Wellcome Trust Sanger Institute, Pfam". Archived from the original on 2014-06-17. Retrieved 2011-05-09.
- ^ "MyHits Dotlet". Retrieved 2011-05-09.
- ^ "Uniprot". Retrieved 2011-05-09.
- ^ a b c d e "SAPS < Sequence Statistics < EMBL-EBI". www.ebi.ac.uk. Retrieved 2019-04-17.
- ^ "DTU Center for Biological Sciences, NetPhos". Retrieved 2011-05-09.
- ^ "DTU Center for Biological Sciences, NetNGlyc". Retrieved 2011-05-09.
- ^ "PHYRE2 Protein Fold Recognition Server". www.sbg.bio.ic.ac.uk. Retrieved 2019-04-20.
- ^ "mentha: the interactome browser". mentha.uniroma2.it. Retrieved 2019-04-20.
- ^ a b www.ebi.ac.uk https://www.ebi.ac.uk/intact/. Retrieved 2019-04-20.
{{cite web}}: Missing or empty|title=(help) - ^ a b "NCBI BLAST". Retrieved 2011-05-09.

