Template talk:Tryptophan metabolism by human microbiota: Difference between revisions
Appearance
Content deleted Content added
Citation bot (talk | contribs) Add: doi-access, bibcode. | Use this bot. Report bugs. | Suggested by Headbomb | #UCB_toolbar |
|||
| (10 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
{{talk header}} |
{{talk header}} |
||
{{WPBS| |
|||
{{WikiProjectBannerShell|1= |
|||
{{WikiProject Medicine |class=Template |
{{WikiProject Medicine |class=Template}} |
||
{{WikiProject Microbiology |class=Template |
{{WikiProject Microbiology |class=Template}} |
||
{{WikiProject |
{{WikiProject Molecular Biology|class=Template|MCB=yes }} |
||
{{WikiProject Pharmacology |class=Template}} |
|||
}} |
}} |
||
==Svg version== |
==Svg version== |
||
{{Ping|Slashme}} Thanks for creating an svg version of the diagram. I noticed there's some weird text rendering in some places, specifically the underlined letters in the following words in the diagram: <u>Tr</u>yptophan (top), anti<u>ox</u>idant (bottom left), <u>fib</u>ril (bottom left), cal<u>ci</u>fication (right). Can you reupload the diagram with a different text font or just tweak the text spacing in those places? [[User:Seppi333|'''< |
{{Ping|Slashme}} Thanks for creating an svg version of the diagram. I noticed there's some weird text rendering in some places, specifically the underlined letters in the following words in the diagram: <u>Tr</u>yptophan (top), anti<u>ox</u>idant (bottom left), <u>fib</u>ril (bottom left), cal<u>ci</u>fication (right). Can you reupload the diagram with a different text font or just tweak the text spacing in those places? [[User:Seppi333|'''<span style="color:#32CD32;">Seppi</span>''<span style="color:Black;">333</span>''''']] ([[User Talk:Seppi333|Insert '''2¢''']]) 21:32, 1 November 2017 (UTC) |
||
:{{Ping|Seppi333}} That's a [https://commons.wikimedia.org/wiki/Librsvg_bugs known bug in the SVG rendering]: I've already done a bit of tweaking, but you're right - it's not quite right yet. I'll give it another go soon. --[[User:Slashme|Slashme]] ([[User talk:Slashme|talk]]) 21:49, 1 November 2017 (UTC) |
:{{Ping|Seppi333}} That's a [https://commons.wikimedia.org/wiki/Librsvg_bugs known bug in the SVG rendering]: I've already done a bit of tweaking, but you're right - it's not quite right yet. I'll give it another go soon. --[[User:Slashme|Slashme]] ([[User talk:Slashme|talk]]) 21:49, 1 November 2017 (UTC) |
||
::{{Ping|Slashme}} Yeah... I'm familiar with the borked text rendering of text in svg images. {{P|2}} Based upon past experience, copying the entire contents of an svg file into a new file in inkscape and re-saving it over the old file sometimes does the trick. The alternative in this particular case would be to take advantage of the fact that this diagram is displayed in [[template:Annotated image 4|an annotated image template]] by overlaying most of the horizontally-oriented text in the image onto the diagram as wikitext. An example of what this looks like is shown in the collapse tab below (note: I greatly increased the default font size for illustrative purposes); {{tlx|Psychostimulant addiction}} is a good example of a wikitext-annotated svg image that's currently used in various articles. [[User:Seppi333|'''< |
::{{Ping|Slashme}} Yeah... I'm familiar with the borked text rendering of text in svg images. {{P|2}} Based upon past experience, copying the entire contents of an svg file into a new file in inkscape and re-saving it over the old file sometimes does the trick. The alternative in this particular case would be to take advantage of the fact that this diagram is displayed in [[template:Annotated image 4|an annotated image template]] by overlaying most of the horizontally-oriented text in the image onto the diagram as wikitext. An example of what this looks like is shown in the collapse tab below (note: I greatly increased the default font size for illustrative purposes); {{tlx|Psychostimulant addiction}} is a good example of a wikitext-annotated svg image that's currently used in various articles. [[User:Seppi333|'''<span style="color:#32CD32;">Seppi</span>''<span style="color:Black;">333</span>''''']] ([[User Talk:Seppi333|Insert '''2¢''']]) 22:06, 1 November 2017 (UTC) |
||
::: The cause of the issue seems to be that librsvg is not handling small font sizes gracefully: probably a rounding error somewhere. I'll try enlarging the text and then scaling it down: that got me almost all the way the first time around. I just need to fix a few things at [[:af:Triptofaan]] first. --[[User:Slashme|Slashme]] ([[User talk:Slashme|talk]]) 22:17, 1 November 2017 (UTC) |
::: The cause of the issue seems to be that librsvg is not handling small font sizes gracefully: probably a rounding error somewhere. I'll try enlarging the text and then scaling it down: that got me almost all the way the first time around. I just need to fix a few things at [[:af:Triptofaan]] first. --[[User:Slashme|Slashme]] ([[User talk:Slashme|talk]]) 22:17, 1 November 2017 (UTC) |
||
::::If you intend to use this image in [[:af:Triptofaan]], it'd probably be better to just remove the text in the image and annotate it since you don't need to create a new svg file for every different Wikipedia where it will be used. It'll only take me about 15-20 minutes to annotate all of the text in this image back onto it if it's removed. Reusing the image on a different language variant of Wikipedia only requires copying [[template:annotated image 4]] to that Wikipedia and replacing the English text in this template's annotations with text in the appropriate language (e.g., compare [[Template:Catecholamine and trace amine biosynthesis]] and [[sr:Шаблон:Catecholamine and trace amine biosynthesis]]). If you'd prefer that approach, all I'd need you to do is delete the following image text: '''Tryptophan''' (top), '''IPA''' (all occurrences in the image), '''I3A''' (all occurences in the image), '''Indole''' (all occurences in the image), '''PXR''' (center-left), '''AhR''' (center-bottom), '''L-cell''' (center), '''TJ''' (center), '''GLP-1''' (center), '''Intestinal epithelium''' (left), '''Gut immune cells''' (center-bottom), '''Indoxyl sulfate''' (center-right), '''AST-120''' (i.e., the red text), the "'''Associated with vascular disease'''" text and all of the text beneath it in the bottom right region of the diagram, the "'''Neuroprotectant'''" text and all of the text beneath it, the "'''Maintains mucosal reactivity'''" text and the text beneath it, the "'''Mucosal homeostasis'''" text and all of the text beneath it. If you don't plan on reusing the diagram on another Wikipedia, then there probably isn't any point in replacing the image text with annotations though. [[User:Seppi333|'''< |
::::If you intend to use this image in [[:af:Triptofaan]], it'd probably be better to just remove the text in the image and annotate it since you don't need to create a new svg file for every different Wikipedia where it will be used. It'll only take me about 15-20 minutes to annotate all of the text in this image back onto it if it's removed. Reusing the image on a different language variant of Wikipedia only requires copying [[template:annotated image 4]] to that Wikipedia and replacing the English text in this template's annotations with text in the appropriate language (e.g., compare [[Template:Catecholamine and trace amine biosynthesis]] and [[sr:Шаблон:Catecholamine and trace amine biosynthesis]]). If you'd prefer that approach, all I'd need you to do is delete the following image text: '''Tryptophan''' (top), '''IPA''' (all occurrences in the image), '''I3A''' (all occurences in the image), '''Indole''' (all occurences in the image), '''PXR''' (center-left), '''AhR''' (center-bottom), '''L-cell''' (center), '''TJ''' (center), '''GLP-1''' (center), '''Intestinal epithelium''' (left), '''Gut immune cells''' (center-bottom), '''Indoxyl sulfate''' (center-right), '''AST-120''' (i.e., the red text), the "'''Associated with vascular disease'''" text and all of the text beneath it in the bottom right region of the diagram, the "'''Neuroprotectant'''" text and all of the text beneath it, the "'''Maintains mucosal reactivity'''" text and the text beneath it, the "'''Mucosal homeostasis'''" text and all of the text beneath it. If you don't plan on reusing the diagram on another Wikipedia, then there probably isn't any point in replacing the image text with annotations though. [[User:Seppi333|'''<span style="color:#32CD32;">Seppi</span>''<span style="color:Black;">333</span>''''']] ([[User Talk:Seppi333|Insert '''2¢''']]) 22:38, 1 November 2017 (UTC) |
||
:::::{{Ping|Seppi333}} Thank you very much for that offer! I've created [[:File:Microbiota-derived 3-Indolepropionic acid-notext.svg|a new file]] with no text. Let me know if you need me to put back the text on the arrows coming down from Tryptophan. --[[User:Slashme|Slashme]] ([[User talk:Slashme|talk]]) 23:10, 1 November 2017 (UTC) |
:::::{{Ping|Seppi333}} Thank you very much for that offer! I've created [[:File:Microbiota-derived 3-Indolepropionic acid-notext.svg|a new file]] with no text. Let me know if you need me to put back the text on the arrows coming down from Tryptophan. --[[User:Slashme|Slashme]] ([[User talk:Slashme|talk]]) 23:10, 1 November 2017 (UTC) |
||
::::::{{Ping|Slashme}} Would you be able to move the circle in the center on the right side (i.e., using the [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4840492/figure/Fig1/ original image] as a reference, the circle with the letters "TJ" inside) into the line that it's next to? "TJ" indicates a [[tight junction]], which is actually what each vertical line in that part of the diagram represents. Alternatively, I suppose the circle could just be removed and the letters "TJ" could just be placed right alongside the line.<br />Anyway, I'm going to finish adding the coordinates for the annotations later since I need to log off for now. It shouldn't take long, so it'll definitely be done by tomorrow. [[User:Seppi333|'''< |
::::::{{Ping|Slashme}} Would you be able to move the circle in the center on the right side (i.e., using the [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4840492/figure/Fig1/ original image] as a reference, the circle with the letters "TJ" inside) into the line that it's next to? "TJ" indicates a [[tight junction]], which is actually what each vertical line in that part of the diagram represents. Alternatively, I suppose the circle could just be removed and the letters "TJ" could just be placed right alongside the line.<br />Anyway, I'm going to finish adding the coordinates for the annotations later since I need to log off for now. It shouldn't take long, so it'll definitely be done by tomorrow. [[User:Seppi333|'''<span style="color:#32CD32;">Seppi</span>''<span style="color:Black;">333</span>''''']] ([[User Talk:Seppi333|Insert '''2¢''']]) 00:25, 2 November 2017 (UTC) |
||
:::::::{{Ping|Seppi333}} Sure thing! I'll do it as soon as I get home this afternoon. By the way, the annotated diagram looks brilliant! --[[User:Slashme|Slashme]] ([[User talk:Slashme|talk]]) 08:25, 2 November 2017 (UTC) |
:::::::{{Ping|Seppi333}} Sure thing! I'll do it as soon as I get home this afternoon. By the way, the annotated diagram looks brilliant! --[[User:Slashme|Slashme]] ([[User talk:Slashme|talk]]) 08:25, 2 November 2017 (UTC) |
||
:::::::{{Ping|Seppi333}} Done - you might need to shift-reload if it still looks as if the circle is in the same place. --[[User:Slashme|Slashme]] ([[User talk:Slashme|talk]]) 19:00, 2 November 2017 (UTC) |
:::::::{{Ping|Seppi333}} Done - you might need to shift-reload if it still looks as if the circle is in the same place. --[[User:Slashme|Slashme]] ([[User talk:Slashme|talk]]) 19:00, 2 November 2017 (UTC) |
||
::::::::Hah, ok. Looks like I need to shift all of the annotations a few pixels to the right. I can get to that in a little bit. [[User:Seppi333|'''< |
::::::::Hah, ok. Looks like I need to shift all of the annotations a few pixels to the right. I can get to that in a little bit. [[User:Seppi333|'''<span style="color:#32CD32;">Seppi</span>''<span style="color:Black;">333</span>''''']] ([[User Talk:Seppi333|Insert '''2¢''']]) 19:02, 2 November 2017 (UTC) |
||
Your waffles are extremely tasty! See how nice [[:af:Triptofaan]] looks now. --[[User:Slashme|Slashme]] ([[User talk:Slashme|talk]]) 21:55, 2 November 2017 (UTC) |
{{od}} Your waffles are extremely tasty! See how nice [[:af:Triptofaan]] looks now. --[[User:Slashme|Slashme]] ([[User talk:Slashme|talk]]) 21:55, 2 November 2017 (UTC) |
||
:Looks great! I'm done with the English version. I said that would take me 20-30 minutes, hahahaha. That took me more like 1-2 hours since I'm excessively finicky when it comes to positioning text within small spaces. [[User:Seppi333|'''<span style="color:#32CD32;">Seppi</span>''<span style="color:Black;">333</span>''''']] ([[User Talk:Seppi333|Insert '''2¢''']]) 03:42, 3 November 2017 (UTC) |
|||
{{Hidden |
{{Hidden |
||
| Line 28: | Line 30: | ||
| content ={{Annotated image 4 |
| content ={{Annotated image 4 |
||
| image = Microbiota-derived 3-Indolepropionic acid.svg |
| image = Microbiota-derived 3-Indolepropionic acid.svg |
||
| header = {{{header|Tryptophan metabolism by {{if pagename|Human microbiota=human gastrointestinal microbiota|other=[[human microbiota|human gastrointestinal microbiota]]}} <small>(</small>{{ |
| header = {{{header|Tryptophan metabolism by {{if pagename|Human microbiota=human gastrointestinal microbiota|other=[[human microbiota|human gastrointestinal microbiota]]}} <small>(</small>{{navbar|mini=y|t|e|template=Tryptophan metabolism by human microbiota}}<small>)</small>}}} |
||
| header_align = {{{header align|center}}} |
| header_align = {{{header align|center}}} |
||
| header_background = {{{header background|#F0F8FF}}} |
| header_background = {{{header background|#F0F8FF}}} |
||
| Line 38: | Line 40: | ||
| height = 470 |
| height = 470 |
||
| alt = Tryptophan metabolism diagram |
| alt = Tryptophan metabolism diagram |
||
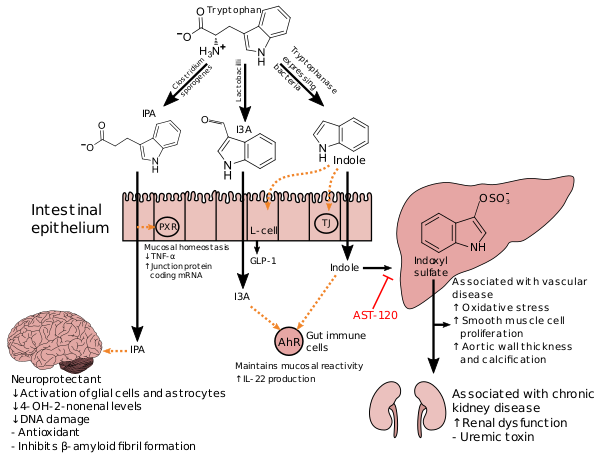
| caption = This diagram shows the biosynthesis of [[bioactive compound]]s ([[indole]] and certain derivatives) from {{if pagename|Tryptophan=tryptophan|other=[[tryptophan]]}} by bacteria in the gut.<ref name="Microbial biosynthesis of bioactive compounds" /> Indole is produced from tryptophan by bacteria that express [[tryptophanase]].<ref name="Microbial biosynthesis of bioactive compounds" /> {{if pagename|Clostridium sporogenes=''Clostridium sporogenes''|other=''[[Clostridium sporogenes]]''}} metabolizes indole into {{if pagename|3-Indolepropionic acid=3-indolepropionic acid|other=[[3-indolepropionic acid]]}} (IPA),<ref name="Microbiome IPA">{{cite journal | vauthors = Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G | title = Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 106 | issue = 10 | pages = 3698–3703 | date = March 2009 | pmid = 19234110 | pmc = 2656143 | doi = 10.1073/pnas.0812874106 | quote = {{if pagename|3-Indolepropionic acid=Production of IPA was shown to be completely dependent on the presence of gut microflora and could be established by colonization with the bacterium Clostridium sporogenes. ... Conversely, a different set of enteric bacteria has been implicated in the metabolic transformation of indole to indole-3-propionic acid (IPA) (27). IPA, also identified only in the plasma of conv mice, has been shown to be a powerful antioxidant (28) ... Although the presence of IPA in mammals has long been ascribed in the literature to bacterial metabolic processes, this conclusion was based on either the production of IPA in ex vivo cultures of individual bacterial species (31) or observed decreases in IPA levels in animals after administration of antibiotics (32). In our own survey of IPA production by representative members of the intestinal flora, only Clostridium sporogenes was found to produce IPA in culture (Table S2). Based on these results, individual GF mice were intentionally colonized with C. sporogenes strain ATCC 15579, and blood samples were taken at several intervals after colonization. IPA was undetectable in the samples taken shortly after introduction of the microbes, and was first observed in the serum 5 days after colonization, reaching plateau values comparable with conv mice by day 10. These colonization studies demonstrate that the introduction of enteric bacteria capable of IPA production in vivo into the gastrointestinal tract is sufficient to introduce IPA into the bloodstream of the host. Also, other GF animals were injected i.p. with either IPA (at 10, 20, or 40 mg/kg) or sterile PBS vehicle, and their serum concentrations of IPA were measured over time. As seen in Table S3, the high serum levels of IPA observed 1 h after injection decreased more than 90% within 5 h, showing that IPA is rapidly cleared from the blood, and that its presence in the serum of conv animals must result from continuous production from 1 or more bacterial species associated with the mammalian gut.|other=Production of IPA was shown to be completely dependent on the presence of gut microflora and could be established by colonization with the bacterium ''Clostridium sporogenes''.}}}}<br />[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2656143/figure/F2/ IPA metabolism diagram]</ref> a highly potent [[neuroprotective]] [[antioxidant]] that scavenges [[hydroxyl radical]]s.<ref name="Microbial biosynthesis of bioactive compounds" /><ref name="Human metabolome IPA">{{cite web|title=3-Indolepropionic acid|website=Human Metabolome Database|publisher=University of Alberta|accessdate=12 October 2015|url=http://www.hmdb.ca/metabolites/HMDB02302|quote=Indole-3-propionate (IPA), a deamination product of tryptophan formed by symbiotic bacteria in the gastrointestinal tract of mammals and birds. 3-Indolepropionic acid has been shown to prevent oxidative stress and death of primary neurons and neuroblastoma cells exposed to the amyloid beta-protein in the form of amyloid fibrils, one of the most prominent neuropathologic features of Alzheimer's disease. 3-Indolepropionic acid also shows a strong level of neuroprotection in two other paradigms of oxidative stress. ({{PMID|10419516}} )<br />Origin: {{bull}} Endogenous {{bull}} Microbial}}</ref><ref name="Indolepropionic acid scavenging">{{cite journal | vauthors = Chyan YJ, Poeggeler B, Omar RA, Chain DG, Frangione B, Ghiso J, Pappolla MA | title = Potent neuroprotective properties against the Alzheimer beta-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid | journal = J. Biol. Chem. | volume = 274 | issue = 31 | pages = 21937–21942 | date = July 1999 | pmid = 10419516 | doi = 10.1074/jbc.274.31.21937| quote = [Indole-3-propionic acid (IPA)] has previously been identified in the plasma and cerebrospinal fluid of humans, but its functions are not known. ... In kinetic competition experiments using free radical-trapping agents, the capacity of IPA to scavenge hydroxyl radicals exceeded that of melatonin, an indoleamine considered to be the most potent naturally occurring scavenger of free radicals. In contrast with other antioxidants, IPA was not converted to reactive intermediates with pro-oxidant activity.}}</ref> In the intestine, IPA binds to [[pregnane X receptor]]s (PXR) in intestinal cells, thereby facilitating mucosal homeostasis and [[Intestinal permeability|barrier function]].<ref name="Microbial biosynthesis of bioactive compounds" /> Following [[Absorption (pharmacokinetics)|absorption]] from the intestine and [[Distribution (pharmacology)|distribution]] to the brain, IPA confers a neuroprotective effect against [[cerebral ischemia]] and [[Alzheimer’s disease]].<ref name="Microbial biosynthesis of bioactive compounds" /> ''[[Lactobacillus]]'' species metabolize tryptophan into [[indole-3-carboxaldehyde|indole-3-aldehyde]] (I3A) which acts on the [[aryl hydrocarbon receptor]] (AhR) in intestinal immune cells, in turn increasing [[interleukin-22]] (IL-22) production.<ref name="Microbial biosynthesis of bioactive compounds" /> Indole itself acts as a [[glucagon-like peptide-1]] (GLP-1) [[secretagogue]] in [[Enteroendocrine_cell#L_cell|intestinal L cells]] and as a ligand for AhR.<ref name="Microbial biosynthesis of bioactive compounds" /> Indole can also be metabolized by the liver into [[indoxyl sulfate]], a compound that is toxic in high concentrations and associated with vascular disease and renal dysfunction.<ref name="Microbial biosynthesis of bioactive compounds" /> AST-120 ([[activated charcoal]]), an intestinal [[sorbent]] that is [[Oral administration|taken by mouth]], [[Adsorption|adsorbs]] indole, in turn decreasing the concentration of indoxyl sulfate in blood plasma.<ref name="Microbial biosynthesis of bioactive compounds">{{cite journal | vauthors = Zhang LS, Davies SS | title = Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions | journal = Genome Med | volume = 8 | issue = 1 | pages = 46 | date = April 2016 | pmid = 27102537 | pmc = 4840492 | doi = 10.1186/s13073-016-0296-x | quote = ''Lactobacillus'' spp. convert tryptophan to indole-3-aldehyde (I3A) through unidentified enzymes [125]. ''Clostridium sporogenes'' convert tryptophan to IPA [6], likely via a tryptophan deaminase. ... IPA also potently scavenges hydroxyl radicals}}<br />[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4840492/table/Tab2/ Table 2: Microbial metabolites: their synthesis, mechanisms of action, and effects on health and disease]<br />[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4840492/figure/Fig1/ Figure 1: Molecular mechanisms of action of indole and its metabolites on host physiology and disease]</ref> |
| caption = This diagram shows the biosynthesis of [[bioactive compound]]s ([[indole]] and certain derivatives) from {{if pagename|Tryptophan=tryptophan|other=[[tryptophan]]}} by bacteria in the gut.<ref name="Microbial biosynthesis of bioactive compounds" /> Indole is produced from tryptophan by bacteria that express [[tryptophanase]].<ref name="Microbial biosynthesis of bioactive compounds" /> {{if pagename|Clostridium sporogenes=''Clostridium sporogenes''|other=''[[Clostridium sporogenes]]''}} metabolizes indole into {{if pagename|3-Indolepropionic acid=3-indolepropionic acid|other=[[3-indolepropionic acid]]}} (IPA),<ref name="Microbiome IPA">{{cite journal | vauthors = Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G | title = Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 106 | issue = 10 | pages = 3698–3703 | date = March 2009 | pmid = 19234110 | pmc = 2656143 | doi = 10.1073/pnas.0812874106 | bibcode = 2009PNAS..106.3698W | quote = {{if pagename|3-Indolepropionic acid=Production of IPA was shown to be completely dependent on the presence of gut microflora and could be established by colonization with the bacterium Clostridium sporogenes. ... Conversely, a different set of enteric bacteria has been implicated in the metabolic transformation of indole to indole-3-propionic acid (IPA) (27). IPA, also identified only in the plasma of conv mice, has been shown to be a powerful antioxidant (28) ... Although the presence of IPA in mammals has long been ascribed in the literature to bacterial metabolic processes, this conclusion was based on either the production of IPA in ex vivo cultures of individual bacterial species (31) or observed decreases in IPA levels in animals after administration of antibiotics (32). In our own survey of IPA production by representative members of the intestinal flora, only Clostridium sporogenes was found to produce IPA in culture (Table S2). Based on these results, individual GF mice were intentionally colonized with C. sporogenes strain ATCC 15579, and blood samples were taken at several intervals after colonization. IPA was undetectable in the samples taken shortly after introduction of the microbes, and was first observed in the serum 5 days after colonization, reaching plateau values comparable with conv mice by day 10. These colonization studies demonstrate that the introduction of enteric bacteria capable of IPA production in vivo into the gastrointestinal tract is sufficient to introduce IPA into the bloodstream of the host. Also, other GF animals were injected i.p. with either IPA (at 10, 20, or 40 mg/kg) or sterile PBS vehicle, and their serum concentrations of IPA were measured over time. As seen in Table S3, the high serum levels of IPA observed 1 h after injection decreased more than 90% within 5 h, showing that IPA is rapidly cleared from the blood, and that its presence in the serum of conv animals must result from continuous production from 1 or more bacterial species associated with the mammalian gut.|other=Production of IPA was shown to be completely dependent on the presence of gut microflora and could be established by colonization with the bacterium ''Clostridium sporogenes''.}} | doi-access = free }}<br />[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2656143/figure/F2/ IPA metabolism diagram]</ref> a highly potent [[neuroprotective]] [[antioxidant]] that scavenges [[hydroxyl radical]]s.<ref name="Microbial biosynthesis of bioactive compounds" /><ref name="Human metabolome IPA">{{cite web|title=3-Indolepropionic acid|website=Human Metabolome Database|publisher=University of Alberta|accessdate=12 October 2015|url=http://www.hmdb.ca/metabolites/HMDB02302|quote=Indole-3-propionate (IPA), a deamination product of tryptophan formed by symbiotic bacteria in the gastrointestinal tract of mammals and birds. 3-Indolepropionic acid has been shown to prevent oxidative stress and death of primary neurons and neuroblastoma cells exposed to the amyloid beta-protein in the form of amyloid fibrils, one of the most prominent neuropathologic features of Alzheimer's disease. 3-Indolepropionic acid also shows a strong level of neuroprotection in two other paradigms of oxidative stress. ({{PMID|10419516}} )<br />Origin: {{bull}} Endogenous {{bull}} Microbial}}</ref><ref name="Indolepropionic acid scavenging">{{cite journal | vauthors = Chyan YJ, Poeggeler B, Omar RA, Chain DG, Frangione B, Ghiso J, Pappolla MA | title = Potent neuroprotective properties against the Alzheimer beta-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid | journal = J. Biol. Chem. | volume = 274 | issue = 31 | pages = 21937–21942 | date = July 1999 | pmid = 10419516 | doi = 10.1074/jbc.274.31.21937| quote = [Indole-3-propionic acid (IPA)] has previously been identified in the plasma and cerebrospinal fluid of humans, but its functions are not known. ... In kinetic competition experiments using free radical-trapping agents, the capacity of IPA to scavenge hydroxyl radicals exceeded that of melatonin, an indoleamine considered to be the most potent naturally occurring scavenger of free radicals. In contrast with other antioxidants, IPA was not converted to reactive intermediates with pro-oxidant activity. | doi-access = free }}</ref> In the intestine, IPA binds to [[pregnane X receptor]]s (PXR) in intestinal cells, thereby facilitating mucosal homeostasis and [[Intestinal permeability|barrier function]].<ref name="Microbial biosynthesis of bioactive compounds" /> Following [[Absorption (pharmacokinetics)|absorption]] from the intestine and [[Distribution (pharmacology)|distribution]] to the brain, IPA confers a neuroprotective effect against [[cerebral ischemia]] and [[Alzheimer’s disease]].<ref name="Microbial biosynthesis of bioactive compounds" /> ''[[Lactobacillus]]'' species metabolize tryptophan into [[indole-3-carboxaldehyde|indole-3-aldehyde]] (I3A) which acts on the [[aryl hydrocarbon receptor]] (AhR) in intestinal immune cells, in turn increasing [[interleukin-22]] (IL-22) production.<ref name="Microbial biosynthesis of bioactive compounds" /> Indole itself acts as a [[glucagon-like peptide-1]] (GLP-1) [[secretagogue]] in [[Enteroendocrine_cell#L_cell|intestinal L cells]] and as a ligand for AhR.<ref name="Microbial biosynthesis of bioactive compounds" /> Indole can also be metabolized by the liver into [[indoxyl sulfate]], a compound that is toxic in high concentrations and associated with vascular disease and renal dysfunction.<ref name="Microbial biosynthesis of bioactive compounds" /> AST-120 ([[activated charcoal]]), an intestinal [[sorbent]] that is [[Oral administration|taken by mouth]], [[Adsorption|adsorbs]] indole, in turn decreasing the concentration of indoxyl sulfate in blood plasma.<ref name="Microbial biosynthesis of bioactive compounds">{{cite journal | vauthors = Zhang LS, Davies SS | title = Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions | journal = Genome Med | volume = 8 | issue = 1 | pages = 46 | date = April 2016 | pmid = 27102537 | pmc = 4840492 | doi = 10.1186/s13073-016-0296-x | quote = ''Lactobacillus'' spp. convert tryptophan to indole-3-aldehyde (I3A) through unidentified enzymes [125]. ''Clostridium sporogenes'' convert tryptophan to IPA [6], likely via a tryptophan deaminase. ... IPA also potently scavenges hydroxyl radicals | doi-access = free }}<br />[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4840492/table/Tab2/ Table 2: Microbial metabolites: their synthesis, mechanisms of action, and effects on health and disease]<br />[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4840492/figure/Fig1/ Figure 1: Molecular mechanisms of action of indole and its metabolites on host physiology and disease]</ref> |
||
| icon = none |
| icon = none |
||
| annotations = |
| annotations = |
||
| Line 48: | Line 50: | ||
}} |
}} |
||
==Template name== |
|||
==Image with annotated text== |
|||
Would this be better named as ..metabolism by [[gut flora]] which is more specific?--[[User:Iztwoz|Iztwoz]] ([[User talk:Iztwoz|talk]]) 07:31, 8 January 2018 (UTC) |
|||
'''To add tomorrow:''' |
|||
*L cell |
|||
*GLP-1 |
|||
*TJ |
|||
:{{Ping|Iztwoz}} When I was creating this template, it didn't occur to me; however, when I was creating the annotated image header, I realized I probably should've named the template "Tryptophan metabolism by human ''gastrointestinal'' microbiota". The use of the term ''flora'' to describe microbes has gradually become more and more archaic in recent decades. I wouldn't mind renaming the template, but a lengthy template name isn't always useful even if it is a better descriptor. Despite that, {{tlx|Tryptophan metabolism by human microbiota}} could always be used to call this template instead of the longer template name since it would become a template redirect if this page is moved to {{tlx|Tryptophan metabolism by human gastrointestinal microbiota}}. So, if you want to rename/move the template to the longer title, I'd be okay with that. [[User:Seppi333|'''<span style="color:#32CD32;">Seppi</span>''<span style="color:Black;">333</span>''''']] ([[User Talk:Seppi333|Insert '''2¢''']]) 00:07, 9 January 2018 (UTC) |
|||
{{Annotated image 4 |
|||
| image = Microbiota-derived 3-Indolepropionic acid-notext.svg |
|||
| header = {{{header|Tryptophan metabolism by {{if pagename|Human microbiota=human gastrointestinal microbiota|other=[[human microbiota|human gastrointestinal microbiota]]}} <small>(</small>{{v|t|e|template=Tryptophan metabolism by human microbiota}}<small>)</small>}}} |
|||
| header_align = {{{header align|center}}} |
|||
| header_background = {{{header background|#F0F8FF}}} |
|||
| align = {{{align|right}}} |
|||
| image-width = 600 |
|||
| image-left = 0 |
|||
| image-top = 10 |
|||
| width = 580 |
|||
| height = 470 |
|||
| alt = Tryptophan metabolism diagram |
|||
| caption = This diagram shows the biosynthesis of [[bioactive compound]]s ([[indole]] and certain derivatives) from {{if pagename|Tryptophan=tryptophan|other=[[tryptophan]]}} by bacteria in the gut.<ref name="Microbial biosynthesis of bioactive compounds" /> Indole is produced from tryptophan by bacteria that express [[tryptophanase]].<ref name="Microbial biosynthesis of bioactive compounds" /> {{if pagename|Clostridium sporogenes=''Clostridium sporogenes''|other=''[[Clostridium sporogenes]]''}} metabolizes indole into {{if pagename|3-Indolepropionic acid=3-indolepropionic acid|other=[[3-indolepropionic acid]]}} (IPA),<ref name="Microbiome IPA">{{cite journal | vauthors = Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G | title = Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 106 | issue = 10 | pages = 3698–3703 | date = March 2009 | pmid = 19234110 | pmc = 2656143 | doi = 10.1073/pnas.0812874106 | quote = {{if pagename|3-Indolepropionic acid=Production of IPA was shown to be completely dependent on the presence of gut microflora and could be established by colonization with the bacterium Clostridium sporogenes. ... Conversely, a different set of enteric bacteria has been implicated in the metabolic transformation of indole to indole-3-propionic acid (IPA) (27). IPA, also identified only in the plasma of conv mice, has been shown to be a powerful antioxidant (28) ... Although the presence of IPA in mammals has long been ascribed in the literature to bacterial metabolic processes, this conclusion was based on either the production of IPA in ex vivo cultures of individual bacterial species (31) or observed decreases in IPA levels in animals after administration of antibiotics (32). In our own survey of IPA production by representative members of the intestinal flora, only Clostridium sporogenes was found to produce IPA in culture (Table S2). Based on these results, individual GF mice were intentionally colonized with C. sporogenes strain ATCC 15579, and blood samples were taken at several intervals after colonization. IPA was undetectable in the samples taken shortly after introduction of the microbes, and was first observed in the serum 5 days after colonization, reaching plateau values comparable with conv mice by day 10. These colonization studies demonstrate that the introduction of enteric bacteria capable of IPA production in vivo into the gastrointestinal tract is sufficient to introduce IPA into the bloodstream of the host. Also, other GF animals were injected i.p. with either IPA (at 10, 20, or 40 mg/kg) or sterile PBS vehicle, and their serum concentrations of IPA were measured over time. As seen in Table S3, the high serum levels of IPA observed 1 h after injection decreased more than 90% within 5 h, showing that IPA is rapidly cleared from the blood, and that its presence in the serum of conv animals must result from continuous production from 1 or more bacterial species associated with the mammalian gut.|other=Production of IPA was shown to be completely dependent on the presence of gut microflora and could be established by colonization with the bacterium ''Clostridium sporogenes''.}}}}<br />[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2656143/figure/F2/ IPA metabolism diagram]</ref> a highly potent [[neuroprotective]] [[antioxidant]] that scavenges [[hydroxyl radical]]s.<ref name="Microbial biosynthesis of bioactive compounds" /><ref name="Human metabolome IPA">{{cite web|title=3-Indolepropionic acid|website=Human Metabolome Database|publisher=University of Alberta|accessdate=12 October 2015|url=http://www.hmdb.ca/metabolites/HMDB02302|quote=Indole-3-propionate (IPA), a deamination product of tryptophan formed by symbiotic bacteria in the gastrointestinal tract of mammals and birds. 3-Indolepropionic acid has been shown to prevent oxidative stress and death of primary neurons and neuroblastoma cells exposed to the amyloid beta-protein in the form of amyloid fibrils, one of the most prominent neuropathologic features of Alzheimer's disease. 3-Indolepropionic acid also shows a strong level of neuroprotection in two other paradigms of oxidative stress. ({{PMID|10419516}} )<br />Origin: {{bull}} Endogenous {{bull}} Microbial}}</ref><ref name="Indolepropionic acid scavenging">{{cite journal | vauthors = Chyan YJ, Poeggeler B, Omar RA, Chain DG, Frangione B, Ghiso J, Pappolla MA | title = Potent neuroprotective properties against the Alzheimer beta-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid | journal = J. Biol. Chem. | volume = 274 | issue = 31 | pages = 21937–21942 | date = July 1999 | pmid = 10419516 | doi = 10.1074/jbc.274.31.21937| quote = [Indole-3-propionic acid (IPA)] has previously been identified in the plasma and cerebrospinal fluid of humans, but its functions are not known. ... In kinetic competition experiments using free radical-trapping agents, the capacity of IPA to scavenge hydroxyl radicals exceeded that of melatonin, an indoleamine considered to be the most potent naturally occurring scavenger of free radicals. In contrast with other antioxidants, IPA was not converted to reactive intermediates with pro-oxidant activity.}}</ref> In the intestine, IPA binds to [[pregnane X receptor]]s (PXR) in intestinal cells, thereby facilitating mucosal homeostasis and [[Intestinal permeability|barrier function]].<ref name="Microbial biosynthesis of bioactive compounds" /> Following [[Absorption (pharmacokinetics)|absorption]] from the intestine and [[Distribution (pharmacology)|distribution]] to the brain, IPA confers a neuroprotective effect against [[cerebral ischemia]] and [[Alzheimer’s disease]].<ref name="Microbial biosynthesis of bioactive compounds" /> ''[[Lactobacillus]]'' species metabolize tryptophan into [[indole-3-carboxaldehyde|indole-3-aldehyde]] (I3A) which acts on the [[aryl hydrocarbon receptor]] (AhR) in intestinal immune cells, in turn increasing [[interleukin-22]] (IL-22) production.<ref name="Microbial biosynthesis of bioactive compounds" /> Indole itself acts as a [[glucagon-like peptide-1]] (GLP-1) [[secretagogue]] in [[Enteroendocrine_cell#L_cell|intestinal L cells]] and as a [[Ligand (biochemistry)|ligand]] for AhR.<ref name="Microbial biosynthesis of bioactive compounds" /> Indole can also be metabolized by the liver into [[indoxyl sulfate]], a compound that is toxic in high concentrations and associated with [[vascular disease]] and [[renal dysfunction]].<ref name="Microbial biosynthesis of bioactive compounds" /> AST-120 ([[activated charcoal]]), an intestinal [[sorbent]] that is [[Oral administration|taken by mouth]], [[Adsorption|adsorbs]] indole, in turn decreasing the concentration of indoxyl sulfate in blood plasma.<ref name="Microbial biosynthesis of bioactive compounds">{{cite journal | vauthors = Zhang LS, Davies SS | title = Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions | journal = Genome Med | volume = 8 | issue = 1 | pages = 46 | date = April 2016 | pmid = 27102537 | pmc = 4840492 | doi = 10.1186/s13073-016-0296-x | quote = ''Lactobacillus'' spp. convert tryptophan to indole-3-aldehyde (I3A) through unidentified enzymes [125]. ''Clostridium sporogenes'' convert tryptophan to IPA [6], likely via a tryptophan deaminase. ... IPA also potently scavenges hydroxyl radicals}}<br />[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4840492/table/Tab2/ Table 2: Microbial metabolites: their synthesis, mechanisms of action, and effects on health and disease]<br />[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4840492/figure/Fig1/ Figure 1: Molecular mechanisms of action of indole and its metabolites on host physiology and disease]</ref> |
|||
| annot-font-size = 14 |
|||
| annot-text-align = left |
|||
| annotations = |
|||
<!--The text annotations are (mostly) sorted into groups by vertical position and then within groups by horizontal position --> |
|||
{{annotation|205|3|[[Tryptophan]]}} |
|||
{{annotation|128|65|[[Clostridium sporogenes|Clostridium<br />sporogenes]]|annot-text-align=center|font-size=12}} |
|||
{{annotation|252|85|[[Lactobacilli|Lacto-<br/>bacilli]]|font-size=12}} |
|||
{{annotation|292|60|[[Tryptophanase]]-<br />expressing<br />bacteria|text-align=center|font-size=12}} |
|||
{{annotation|148|116|[[3-Indolepropionic acid|IPA]]|font-size=14}} |
|||
{{annotation|236|131|I3A|font-size=14}} |
|||
{{annotation|313|115|[[Indole]]|font-size=14}} |
|||
{{annotation|470|174|[[Liver]]|text-align=center|font-size=18}} |
|||
{{annotation|37|301|[[Human brain|Brain]]|font-size=18}} |
|||
{{annotation|133|352|[[3-Indolepropionic acid|IPA]]|font-size=14}} |
|||
{{annotation|236|299|I3A|font-size=14}} |
|||
{{annotation|328|269|[[Indole]]|font-size=14}} |
|||
{{annotation|413|261|Indoxyl<br />sulfate|text-align=center|font-size=12}} |
|||
{{annotation|350|318|[[Activated charcoal|<span style="color:red">AST-120</span>]]|font-size=14}} |
|||
{{annotation|281|346|[[Aryl hydrocarbon receptor|AhR]]|font-size=12}} |
|||
{{annotation|269|297|Intestinal<br />[[immune cells|immune<br />cells]]|text-align=center|font-size=11}} |
|||
{{annotation|40|209|[[Intestinal epithelium|Intestinal<br />epithelium]]|text-align=center|font-size=18}} |
|||
{{annotation|161.5|231.5|[[Pregnane X receptor|PXR]]|font-size=11}} |
|||
{{annotation|144|255|Mucosal homeostasis<br />↓[[Tumor necrosis factor alpha|TNF-α]]<br />↑[[Tight junction|Junction]] protein-<br />coding [[Messenger RNA|mRNA]]s|font-size=10}} |
|||
{{annotation|252.5|237|[[Enteroendocrine cell#L cell|L cell]]|font-size=12}} |
|||
{{annotation|249|265|[[Glucagon-like peptide 1|GLP-1]]|font-size=12}} |
|||
{{annotation|335|227|[[Tight junction|T J]]|font-size=12}} |
|||
{{annotation|15|390|[[Neuroprotectant]]<br />↓Activation of [[glial cell]]s and [[astrocyte]]s<br />↓[[4-Hydroxy-2-nonenal]] levels<br />↓[[DNA]] damage<br />–[[Antioxidant]]<br />–Inhibits [[β-amyloid]] fibril formation|font-size=10}} |
|||
{{annotation|243|370|Maintains mucosal reactivity<br />↑[[Interleukin 22|IL-22]] production|font-size=10}} |
|||
{{annotation|458|290|Associated with [[vascular disease]]<br />↑[[Oxidative stress]]<br />↑[[Smooth muscle cell]] [[Cell proliferation|proliferation]]<br />↑[[Aorta|Aortic wall]] thickness and [[aortic calcification|calcification]]|font-size=10}} |
|||
{{annotation|458|391|Associated with [[chronic kidney disease]]<br />↑[[Renal dysfunction]]<br />–[[Uremic toxin]]|font-size=10}} |
|||
{{annotation|381|448|[[Kidneys]]|font-size=18}} |
|||
}} |
|||
{{cot|reflist}} |
|||
{{reflist}} |
|||
{{cob}} |
|||
Latest revision as of 01:42, 13 December 2023
| This is the talk page for discussing improvements to the Tryptophan metabolism by human microbiota template. |
|
| This template does not require a rating on Wikipedia's content assessment scale. It is of interest to the following WikiProjects: | ||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||
Svg version
[edit]@Slashme: Thanks for creating an svg version of the diagram. I noticed there's some weird text rendering in some places, specifically the underlined letters in the following words in the diagram: Tryptophan (top), antioxidant (bottom left), fibril (bottom left), calcification (right). Can you reupload the diagram with a different text font or just tweak the text spacing in those places? Seppi333 (Insert 2¢) 21:32, 1 November 2017 (UTC)
- @Seppi333: That's a known bug in the SVG rendering: I've already done a bit of tweaking, but you're right - it's not quite right yet. I'll give it another go soon. --Slashme (talk) 21:49, 1 November 2017 (UTC)
- @Slashme: Yeah... I'm familiar with the borked text rendering of text in svg images.
 Based upon past experience, copying the entire contents of an svg file into a new file in inkscape and re-saving it over the old file sometimes does the trick. The alternative in this particular case would be to take advantage of the fact that this diagram is displayed in an annotated image template by overlaying most of the horizontally-oriented text in the image onto the diagram as wikitext. An example of what this looks like is shown in the collapse tab below (note: I greatly increased the default font size for illustrative purposes);
Based upon past experience, copying the entire contents of an svg file into a new file in inkscape and re-saving it over the old file sometimes does the trick. The alternative in this particular case would be to take advantage of the fact that this diagram is displayed in an annotated image template by overlaying most of the horizontally-oriented text in the image onto the diagram as wikitext. An example of what this looks like is shown in the collapse tab below (note: I greatly increased the default font size for illustrative purposes); {{Psychostimulant addiction}}is a good example of a wikitext-annotated svg image that's currently used in various articles. Seppi333 (Insert 2¢) 22:06, 1 November 2017 (UTC)- The cause of the issue seems to be that librsvg is not handling small font sizes gracefully: probably a rounding error somewhere. I'll try enlarging the text and then scaling it down: that got me almost all the way the first time around. I just need to fix a few things at af:Triptofaan first. --Slashme (talk) 22:17, 1 November 2017 (UTC)
- If you intend to use this image in af:Triptofaan, it'd probably be better to just remove the text in the image and annotate it since you don't need to create a new svg file for every different Wikipedia where it will be used. It'll only take me about 15-20 minutes to annotate all of the text in this image back onto it if it's removed. Reusing the image on a different language variant of Wikipedia only requires copying template:annotated image 4 to that Wikipedia and replacing the English text in this template's annotations with text in the appropriate language (e.g., compare Template:Catecholamine and trace amine biosynthesis and sr:Шаблон:Catecholamine and trace amine biosynthesis). If you'd prefer that approach, all I'd need you to do is delete the following image text: Tryptophan (top), IPA (all occurrences in the image), I3A (all occurences in the image), Indole (all occurences in the image), PXR (center-left), AhR (center-bottom), L-cell (center), TJ (center), GLP-1 (center), Intestinal epithelium (left), Gut immune cells (center-bottom), Indoxyl sulfate (center-right), AST-120 (i.e., the red text), the "Associated with vascular disease" text and all of the text beneath it in the bottom right region of the diagram, the "Neuroprotectant" text and all of the text beneath it, the "Maintains mucosal reactivity" text and the text beneath it, the "Mucosal homeostasis" text and all of the text beneath it. If you don't plan on reusing the diagram on another Wikipedia, then there probably isn't any point in replacing the image text with annotations though. Seppi333 (Insert 2¢) 22:38, 1 November 2017 (UTC)
- @Seppi333: Thank you very much for that offer! I've created a new file with no text. Let me know if you need me to put back the text on the arrows coming down from Tryptophan. --Slashme (talk) 23:10, 1 November 2017 (UTC)
- @Slashme: Would you be able to move the circle in the center on the right side (i.e., using the original image as a reference, the circle with the letters "TJ" inside) into the line that it's next to? "TJ" indicates a tight junction, which is actually what each vertical line in that part of the diagram represents. Alternatively, I suppose the circle could just be removed and the letters "TJ" could just be placed right alongside the line.
Anyway, I'm going to finish adding the coordinates for the annotations later since I need to log off for now. It shouldn't take long, so it'll definitely be done by tomorrow. Seppi333 (Insert 2¢) 00:25, 2 November 2017 (UTC)- @Seppi333: Sure thing! I'll do it as soon as I get home this afternoon. By the way, the annotated diagram looks brilliant! --Slashme (talk) 08:25, 2 November 2017 (UTC)
- @Seppi333: Done - you might need to shift-reload if it still looks as if the circle is in the same place. --Slashme (talk) 19:00, 2 November 2017 (UTC)
- Hah, ok. Looks like I need to shift all of the annotations a few pixels to the right. I can get to that in a little bit. Seppi333 (Insert 2¢) 19:02, 2 November 2017 (UTC)
- @Slashme: Would you be able to move the circle in the center on the right side (i.e., using the original image as a reference, the circle with the letters "TJ" inside) into the line that it's next to? "TJ" indicates a tight junction, which is actually what each vertical line in that part of the diagram represents. Alternatively, I suppose the circle could just be removed and the letters "TJ" could just be placed right alongside the line.
- @Seppi333: Thank you very much for that offer! I've created a new file with no text. Let me know if you need me to put back the text on the arrows coming down from Tryptophan. --Slashme (talk) 23:10, 1 November 2017 (UTC)
- If you intend to use this image in af:Triptofaan, it'd probably be better to just remove the text in the image and annotate it since you don't need to create a new svg file for every different Wikipedia where it will be used. It'll only take me about 15-20 minutes to annotate all of the text in this image back onto it if it's removed. Reusing the image on a different language variant of Wikipedia only requires copying template:annotated image 4 to that Wikipedia and replacing the English text in this template's annotations with text in the appropriate language (e.g., compare Template:Catecholamine and trace amine biosynthesis and sr:Шаблон:Catecholamine and trace amine biosynthesis). If you'd prefer that approach, all I'd need you to do is delete the following image text: Tryptophan (top), IPA (all occurrences in the image), I3A (all occurences in the image), Indole (all occurences in the image), PXR (center-left), AhR (center-bottom), L-cell (center), TJ (center), GLP-1 (center), Intestinal epithelium (left), Gut immune cells (center-bottom), Indoxyl sulfate (center-right), AST-120 (i.e., the red text), the "Associated with vascular disease" text and all of the text beneath it in the bottom right region of the diagram, the "Neuroprotectant" text and all of the text beneath it, the "Maintains mucosal reactivity" text and the text beneath it, the "Mucosal homeostasis" text and all of the text beneath it. If you don't plan on reusing the diagram on another Wikipedia, then there probably isn't any point in replacing the image text with annotations though. Seppi333 (Insert 2¢) 22:38, 1 November 2017 (UTC)
- The cause of the issue seems to be that librsvg is not handling small font sizes gracefully: probably a rounding error somewhere. I'll try enlarging the text and then scaling it down: that got me almost all the way the first time around. I just need to fix a few things at af:Triptofaan first. --Slashme (talk) 22:17, 1 November 2017 (UTC)
- @Slashme: Yeah... I'm familiar with the borked text rendering of text in svg images.
Your waffles are extremely tasty! See how nice af:Triptofaan looks now. --Slashme (talk) 21:55, 2 November 2017 (UTC)
- Looks great! I'm done with the English version. I said that would take me 20-30 minutes, hahahaha. That took me more like 1-2 hours since I'm excessively finicky when it comes to positioning text within small spaces. Seppi333 (Insert 2¢) 03:42, 3 November 2017 (UTC)
An example of this template with an annotation
Tryptophan metabolism by human gastrointestinal microbiota ()
This diagram shows the biosynthesis of bioactive compounds (indole and certain derivatives) from tryptophan by bacteria in the gut.[1] Indole is produced from tryptophan by bacteria that express tryptophanase.[1] Clostridium sporogenes metabolizes indole into 3-indolepropionic acid (IPA),[2] a highly potent neuroprotective antioxidant that scavenges hydroxyl radicals.[1][3][4] In the intestine, IPA binds to pregnane X receptors (PXR) in intestinal cells, thereby facilitating mucosal homeostasis and barrier function.[1] Following absorption from the intestine and distribution to the brain, IPA confers a neuroprotective effect against cerebral ischemia and Alzheimer’s disease.[1] Lactobacillus species metabolize tryptophan into indole-3-aldehyde (I3A) which acts on the aryl hydrocarbon receptor (AhR) in intestinal immune cells, in turn increasing interleukin-22 (IL-22) production.[1] Indole itself acts as a glucagon-like peptide-1 (GLP-1) secretagogue in intestinal L cells and as a ligand for AhR.[1] Indole can also be metabolized by the liver into indoxyl sulfate, a compound that is toxic in high concentrations and associated with vascular disease and renal dysfunction.[1] AST-120 (activated charcoal), an intestinal sorbent that is taken by mouth, adsorbs indole, in turn decreasing the concentration of indoxyl sulfate in blood plasma.[1]
|
reflist
|
|---|
|
Template name
[edit]Would this be better named as ..metabolism by gut flora which is more specific?--Iztwoz (talk) 07:31, 8 January 2018 (UTC)
- @Iztwoz: When I was creating this template, it didn't occur to me; however, when I was creating the annotated image header, I realized I probably should've named the template "Tryptophan metabolism by human gastrointestinal microbiota". The use of the term flora to describe microbes has gradually become more and more archaic in recent decades. I wouldn't mind renaming the template, but a lengthy template name isn't always useful even if it is a better descriptor. Despite that,
{{Tryptophan metabolism by human microbiota}}could always be used to call this template instead of the longer template name since it would become a template redirect if this page is moved to{{Tryptophan metabolism by human gastrointestinal microbiota}}. So, if you want to rename/move the template to the longer title, I'd be okay with that. Seppi333 (Insert 2¢) 00:07, 9 January 2018 (UTC)
Categories:
- Template-Class medicine articles
- NA-importance medicine articles
- All WikiProject Medicine pages
- Template-Class Microbiology articles
- NA-importance Microbiology articles
- WikiProject Microbiology articles
- Template-Class Molecular Biology articles
- NA-importance Molecular Biology articles
- Template-Class MCB articles
- NA-importance MCB articles
- WikiProject Molecular and Cellular Biology articles
- All WikiProject Molecular Biology pages
- Template-Class pharmacology articles
- NA-importance pharmacology articles
- WikiProject Pharmacology articles