Fuzzy clustering: Difference between revisions
Rescuing 1 sources and tagging 0 as dead.) #IABot (v2.0.9.5 |
|||
| (21 intermediate revisions by 14 users not shown) | |||
| Line 1: | Line 1: | ||
{{Short description|Type of clustering of data points}} |
|||
{{machine learning bar}} |
{{machine learning bar}} |
||
'''Fuzzy clustering''' (also referred to as '''soft clustering''' or '''soft ''k''-means''') is a form of clustering in which each [[data point]] can belong to more than one cluster. |
'''Fuzzy clustering''' (also referred to as '''soft clustering''' or '''soft ''k''-means''') is a form of clustering in which each [[data point]] can belong to more than one cluster. |
||
| Line 5: | Line 6: | ||
== Comparison to hard clustering == |
== Comparison to hard clustering == |
||
In non-fuzzy clustering (also known as hard clustering), data |
In non-fuzzy clustering (also known as hard clustering), data are divided into distinct clusters, where each data point can only belong to exactly one cluster. In fuzzy clustering, data points can potentially belong to multiple clusters. For example, an apple can be red or green (hard clustering), but an apple can also be red AND green (fuzzy clustering). Here, the apple can be red to a certain degree as well as green to a certain degree. Instead of the apple belonging to green [green = 1] and not red [red = 0], the apple can belong to green [green = 0.5] and red [red = 0.5]. These value are normalized between 0 and 1; however, they do not represent probabilities, so the two values do not need to add up to 1. |
||
== Membership == |
== Membership == |
||
| Line 46: | Line 47: | ||
The FCM aims to minimize an objective function: |
The FCM aims to minimize an objective function: |
||
:<math> |
:<math>J(W,C) = \sum_{i=1}^{n} \sum_{j=1}^{c} w_{ij}^m \left\|\mathbf{x}_i - \mathbf{c}_j \right\|^2</math>, |
||
where: |
where: |
||
:<math>w_{ij} = \frac{1}{\sum_{k=1}^{c} \left(\frac{\left\|\mathbf{x}_i - \mathbf{c}_j \right\|}{\left\|\mathbf{x}_i - \mathbf{c}_k \right\|}\right)^{\frac{2}{m-1}}} |
:<math>w_{ij} = \frac{1}{\sum_{k=1}^{c} \left(\frac{\left\|\mathbf{x}_i - \mathbf{c}_j \right\|}{\left\|\mathbf{x}_i - \mathbf{c}_k \right\|}\right)^{\frac{2}{m-1}}}</math>. |
||
=== Comparison to K-means clustering === |
=== Comparison to K-means clustering === |
||
K-means clustering also attempts to minimize the objective function shown above |
K-means clustering also attempts to minimize the objective function shown above, except that in K-means, the membership values are either zero or one, and cannot take values in between, i.e. <math> w_{ij} \in \{0,1\} </math>. In Fuzzy C-means, the degree of fuzziness is parametrized by <math> m \in (1, \infty )</math>, where a larger <math> m </math> results in fuzzier clusters. In the limit <math> m \rightarrow 1</math>, the memberships, <math> w_{ij}</math> , converge to 0 or 1, and the Fuzzy C-means objective coincides with that of K-means. In the absence of experimentation or domain knowledge, <math> m </math> is commonly set to 2. The algorithm minimizes intra-cluster variance as well, but has the same problems as 'k'-means; the minimum is a local minimum, and the results depend on the initial choice of weights. |
||
=== Implementation === |
|||
There are several implementations of this algorithm that are publicly available.<ref>{{Citation |last=Alobaid |first=Ahmad |title=fuzzycmeans: Fuzzy c-means according to the research paper by James C. Bezdek et. al |url=https://github.com/oeg-upm/fuzzy-c-means |access-date=2023-01-18}}</ref><ref>{{Citation |last=Dias |first=Madson |title=fuzzy-c-means: A simple python implementation of Fuzzy C-means algorithm. |url=https://github.com/omadson/fuzzy-c-means |access-date=2023-01-18}}</ref> |
|||
== Related algorithms == |
== Related algorithms == |
||
| Line 70: | Line 75: | ||
== Applications == |
== Applications == |
||
Clustering problems have applications in surface science, biology, medicine, psychology, economics, and many other disciplines.<ref name=":0">{{Cite journal| |
Clustering problems have applications in surface science, biology, medicine, psychology, economics, and many other disciplines.<ref name=":0">{{Cite journal|last1=Ben-Dor|first1=Amir|last2=Shamir|first2=Ron|last3=Yakhini|first3=Zohar|date=1999-10-01|title=Clustering Gene Expression Patterns|journal=Journal of Computational Biology|volume=6|issue=3–4|pages=281–297|doi=10.1089/106652799318274|issn=1066-5277|pmid=10582567|citeseerx=10.1.1.34.5341}}</ref> |
||
=== Bioinformatics === |
=== Bioinformatics === |
||
In the field of bioinformatics, clustering is used for a number of applications. One use is as a [[pattern recognition]] technique to analyze gene expression data from |
In the field of bioinformatics, clustering is used for a number of applications. One use is as a [[pattern recognition]] technique to analyze gene expression data from RNA-sequencing data or other technologies.<ref>{{Cite journal|last=Valafar|first=Faramarz|date=2002-12-01|title=Pattern Recognition Techniques in Microarray Data Analysis|journal=Annals of the New York Academy of Sciences|language=en|volume=980|issue=1|pages=41–64|doi=10.1111/j.1749-6632.2002.tb04888.x|pmid=12594081|bibcode=2002NYASA.980...41V |issn=1749-6632|citeseerx=10.1.1.199.6445|s2cid=343093 }}</ref> In this case, genes with similar expression patterns are grouped into the same cluster, and different clusters display distinct, well-separated patterns of expression. Use of clustering can provide insight into gene function and regulation.<ref name=":0" /> Because fuzzy clustering allows genes to belong to more than one cluster, it allows for the identification of genes that are conditionally co-regulated or co-expressed.<ref>Valafar F. Pattern recognition techniques in microarray data analysis. Annals of the New York Academy of Sciences. 2002 Dec 1;980(1):41-64.</ref> For example, one gene may be acted on by more than one [[transcription factor]], and one gene may encode a protein that has more than one function. Thus, fuzzy clustering is more appropriate than hard clustering. |
||
=== Image analysis === |
=== Image analysis === |
||
Fuzzy c-means has been a very important tool for image processing in clustering objects in an image. In the |
Fuzzy c-means has been a very important tool for image processing in clustering objects in an image. In the 1970s, mathematicians introduced the spatial term into the FCM algorithm to improve the accuracy of clustering under noise.<ref name="fuzzy c means">{{Cite journal|url=http://www.cvip.uofl.edu/wwwcvip/research/publications/Pub_Pdf/2002/3.pdf|title=A Modified Fuzzy C-Means Algorithm for Bias Field Estimation and Segmentation of MRI Data|journal=IEEE Transactions on Medical Imaging|volume=21|issue=3|year=2002|pages=193–199|first1=Mohamed N.|last1=Ahmed|first2=Sameh M.|last2=Yamany|first3=Nevin|last3=Mohamed|first4=Aly A.|last4=Farag|author-link4=Aly Farag|first5=Thomas|last5=Moriarty|doi=10.1109/42.996338|pmid=11989844|citeseerx=10.1.1.331.9742|s2cid=8480349|access-date=2011-10-02|archive-date=2011-10-02|archive-url=https://web.archive.org/web/20111002132049/http://www.cvip.uofl.edu/wwwcvip/research/publications/Pub_Pdf/2002/3.pdf|url-status=dead}}.</ref> Furthermore, FCM algorithms have been used to distinguish between different activities using image-based features such as the Hu and the Zernike Moments.<ref>{{Cite journal|last=Banerjee|first=Tanvi|date=2014|title=Day or Night Activity Recognition From Video Using Fuzzy Clustering Techniques|journal=IEEE Transactions on Fuzzy Systems|volume=22|issue=3|pages=483–493|doi=10.1109/TFUZZ.2013.2260756|citeseerx=10.1.1.652.2819|s2cid=11606344 }}</ref> Alternatively, A [[fuzzy logic]] model can be described on [[fuzzy set]]s that are defined on three components of the HSL color space [[HSL and HSV]]; The membership functions aim to describe colors follow the human intuition of color identification.<ref name="fuzzyset">{{Cite book|journal=Robocup|volume=5001|year=2008|pages= 548–555|first1=Kashani|last1=Alireza|first2=Amir|last2=Kashani|first3= Nargess |last3=Milani|first4=Peyman|last4=Akhlaghi|first5=Kaveh|last5=Khezri|title=RoboCup 2007: Robot Soccer World Cup XI |chapter=Robust Color Classification Using Fuzzy Reasoning and Genetic Algorithms in RoboCup Soccer Leagues |doi=10.1007/978-3-540-68847-1_59|series = Lecture Notes in Computer Science|isbn = 978-3-540-68846-4}}</ref> |
||
=== Marketing === |
=== Marketing === |
||
| Line 86: | Line 91: | ||
== Image processing example == |
== Image processing example == |
||
[[File:Fuzzy Clustering example.jpg|thumb|168x168px|Image segmented by fuzzy clustering, with the original (top left), clustered (top right), and membership map (bottom)]] |
[[File:Fuzzy Clustering example.jpg|thumb|168x168px|Image segmented by fuzzy clustering, with the original (top left), clustered (top right), and membership map (bottom)]] |
||
[[Image segmentation]] using [[k-means clustering]] algorithms has long been used for pattern recognition, object detection, and medical imaging. However, due to real world limitations such as noise, shadowing, and variations in cameras, traditional hard clustering is often unable to reliably perform image processing tasks as stated above. |
[[Image segmentation]] using [[k-means clustering]] algorithms has long been used for pattern recognition, object detection, and medical imaging. However, due to real world limitations such as noise, shadowing, and variations in cameras, traditional hard clustering is often unable to reliably perform image processing tasks as stated above.{{cn|date=December 2021}} Fuzzy clustering has been proposed as a more applicable algorithm in the performance to these tasks. Given is gray scale image that has undergone fuzzy clustering in Matlab.<ref>{{Cite web|url=https://www.mathworks.com/help/fuzzy/fuzzy-clustering.html|title=Fuzzy Clustering - MATLAB & Simulink|website=www.mathworks.com|access-date=2017-05-03}}</ref> The original image is seen next to a clustered image. Colors are used to give a visual representation of the three distinct clusters used to identify the membership of each pixel. Below, a chart is given that defines the fuzzy membership coefficients of their corresponding intensity values. |
||
Depending on the application for which the fuzzy clustering coefficients are to be used, different pre-processing techniques can be applied to [[RGB color space|RGB]] images. RGB to [[HCL color space|HCL]] conversion is common practice.<ref>{{Cite book|title=Systemic Approaches in Bioinformatics and Computational Systems Biology|last=Lecca|first=Paola|publisher=IGI Global|year=2011|isbn=9781613504369|pages=9}}</ref> |
Depending on the application for which the fuzzy clustering coefficients are to be used, different pre-processing techniques can be applied to [[RGB color space|RGB]] images. RGB to [[HCL color space|HCL]] conversion is common practice.<ref>{{Cite book|title=Systemic Approaches in Bioinformatics and Computational Systems Biology|last=Lecca|first=Paola|publisher=IGI Global|year=2011|isbn=9781613504369|pages=9}}</ref> |
||
Latest revision as of 11:51, 15 May 2024
| Part of a series on |
| Machine learning and data mining |
|---|
Fuzzy clustering (also referred to as soft clustering or soft k-means) is a form of clustering in which each data point can belong to more than one cluster.
Clustering or cluster analysis involves assigning data points to clusters such that items in the same cluster are as similar as possible, while items belonging to different clusters are as dissimilar as possible. Clusters are identified via similarity measures. These similarity measures include distance, connectivity, and intensity. Different similarity measures may be chosen based on the data or the application.[1]
Comparison to hard clustering
[edit]In non-fuzzy clustering (also known as hard clustering), data are divided into distinct clusters, where each data point can only belong to exactly one cluster. In fuzzy clustering, data points can potentially belong to multiple clusters. For example, an apple can be red or green (hard clustering), but an apple can also be red AND green (fuzzy clustering). Here, the apple can be red to a certain degree as well as green to a certain degree. Instead of the apple belonging to green [green = 1] and not red [red = 0], the apple can belong to green [green = 0.5] and red [red = 0.5]. These value are normalized between 0 and 1; however, they do not represent probabilities, so the two values do not need to add up to 1.
Membership
[edit]Membership grades are assigned to each of the data points (tags). These membership grades indicate the degree to which data points belong to each cluster. Thus, points on the edge of a cluster, with lower membership grades, may be in the cluster to a lesser degree than points in the center of cluster.
Fuzzy C-means clustering
[edit]One of the most widely used fuzzy clustering algorithms is the Fuzzy C-means clustering (FCM) algorithm.
History
[edit]Fuzzy c-means (FCM) clustering was developed by J.C. Dunn in 1973,[2] and improved by J.C. Bezdek in 1981.[3]
General description
[edit]The fuzzy c-means algorithm is very similar to the k-means algorithm:
- Choose a number of clusters.
- Assign coefficients randomly to each data point for being in the clusters.
- Repeat until the algorithm has converged (that is, the coefficients' change between two iterations is no more than , the given sensitivity threshold) :
- Compute the centroid for each cluster (shown below).
- For each data point, compute its coefficients of being in the clusters.
Centroid
[edit]Any point x has a set of coefficients giving the degree of being in the kth cluster wk(x). With fuzzy c-means, the centroid of a cluster is the mean of all points, weighted by their degree of belonging to the cluster, or, mathematically,
where m is the hyper- parameter that controls how fuzzy the cluster will be. The higher it is, the fuzzier the cluster will be in the end.
Algorithm
[edit]The FCM algorithm attempts to partition a finite collection of elements into a collection of c fuzzy clusters with respect to some given criterion.
Given a finite set of data, the algorithm returns a list of cluster centres and a partition matrix
, where each element, , tells the degree to which element, , belongs to cluster .
The FCM aims to minimize an objective function:
- ,
where:
- .
Comparison to K-means clustering
[edit]K-means clustering also attempts to minimize the objective function shown above, except that in K-means, the membership values are either zero or one, and cannot take values in between, i.e. . In Fuzzy C-means, the degree of fuzziness is parametrized by , where a larger results in fuzzier clusters. In the limit , the memberships, , converge to 0 or 1, and the Fuzzy C-means objective coincides with that of K-means. In the absence of experimentation or domain knowledge, is commonly set to 2. The algorithm minimizes intra-cluster variance as well, but has the same problems as 'k'-means; the minimum is a local minimum, and the results depend on the initial choice of weights.
Implementation
[edit]There are several implementations of this algorithm that are publicly available.[4][5]
Related algorithms
[edit]Fuzzy C-means (FCM) with automatically determined for the number of clusters could enhance the detection accuracy.[6] Using a mixture of Gaussians along with the expectation-maximization algorithm is a more statistically formalized method which includes some of these ideas: partial membership in classes.
Example
[edit]To better understand this principle, a classic example of mono-dimensional data is given below on an x axis.

This data set can be traditionally grouped into two clusters. By selecting a threshold on the x-axis, the data is separated into two clusters. The resulting clusters are labelled 'A' and 'B', as seen in the following image. Each point belonging to the data set would therefore have a membership coefficient of 1 or 0. This membership coefficient of each corresponding data point is represented by the inclusion of the y-axis.

In fuzzy clustering, each data point can have membership to multiple clusters. By relaxing the definition of membership coefficients from strictly 1 or 0, these values can range from any value from 1 to 0. The following image shows the data set from the previous clustering, but now fuzzy c-means clustering is applied. First, a new threshold value defining two clusters may be generated. Next, new membership coefficients for each data point are generated based on clusters centroids, as well as distance from each cluster centroid.
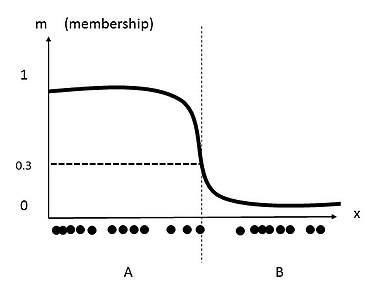
As one can see, the middle data point belongs to cluster A and cluster B. the value of 0.3 is this data point's membership coefficient for cluster A .[7]
Applications
[edit]Clustering problems have applications in surface science, biology, medicine, psychology, economics, and many other disciplines.[8]
Bioinformatics
[edit]In the field of bioinformatics, clustering is used for a number of applications. One use is as a pattern recognition technique to analyze gene expression data from RNA-sequencing data or other technologies.[9] In this case, genes with similar expression patterns are grouped into the same cluster, and different clusters display distinct, well-separated patterns of expression. Use of clustering can provide insight into gene function and regulation.[8] Because fuzzy clustering allows genes to belong to more than one cluster, it allows for the identification of genes that are conditionally co-regulated or co-expressed.[10] For example, one gene may be acted on by more than one transcription factor, and one gene may encode a protein that has more than one function. Thus, fuzzy clustering is more appropriate than hard clustering.
Image analysis
[edit]Fuzzy c-means has been a very important tool for image processing in clustering objects in an image. In the 1970s, mathematicians introduced the spatial term into the FCM algorithm to improve the accuracy of clustering under noise.[11] Furthermore, FCM algorithms have been used to distinguish between different activities using image-based features such as the Hu and the Zernike Moments.[12] Alternatively, A fuzzy logic model can be described on fuzzy sets that are defined on three components of the HSL color space HSL and HSV; The membership functions aim to describe colors follow the human intuition of color identification.[13]
Marketing
[edit]In marketing, customers can be grouped into fuzzy clusters based on their needs, brand choices, psycho-graphic profiles, or other marketing related partitions.[citation needed]
Image processing example
[edit]
Image segmentation using k-means clustering algorithms has long been used for pattern recognition, object detection, and medical imaging. However, due to real world limitations such as noise, shadowing, and variations in cameras, traditional hard clustering is often unable to reliably perform image processing tasks as stated above.[citation needed] Fuzzy clustering has been proposed as a more applicable algorithm in the performance to these tasks. Given is gray scale image that has undergone fuzzy clustering in Matlab.[14] The original image is seen next to a clustered image. Colors are used to give a visual representation of the three distinct clusters used to identify the membership of each pixel. Below, a chart is given that defines the fuzzy membership coefficients of their corresponding intensity values.
Depending on the application for which the fuzzy clustering coefficients are to be used, different pre-processing techniques can be applied to RGB images. RGB to HCL conversion is common practice.[15]
See also
[edit]- FLAME Clustering
- Cluster Analysis
- Expectation-maximization algorithm (a similar, but more statistically formalized method)
References
[edit]- ^ "Fuzzy Clustering". reference.wolfram.com. Retrieved 2016-04-26.
- ^ Dunn, J. C. (1973-01-01). "A Fuzzy Relative of the ISODATA Process and Its Use in Detecting Compact Well-Separated Clusters". Journal of Cybernetics. 3 (3): 32–57. doi:10.1080/01969727308546046. ISSN 0022-0280.
- ^ Bezdek, James C. (1981). Pattern Recognition with Fuzzy Objective Function Algorithms. ISBN 0-306-40671-3.
- ^ Alobaid, Ahmad, fuzzycmeans: Fuzzy c-means according to the research paper by James C. Bezdek et. al, retrieved 2023-01-18
- ^ Dias, Madson, fuzzy-c-means: A simple python implementation of Fuzzy C-means algorithm., retrieved 2023-01-18
- ^ Said, E El-Khamy; Rowayda A Sadek; Mohamed A El-Khoreby (October 2015). "An efficient brain mass detection with adaptive clustered based fuzzy C-mean and thresholding". 2015 IEEE International Conference on Signal and Image Processing Applications (ICSIPA): 429–433.
- ^ "Clustering - Fuzzy C-means". home.deib.polimi.it. Retrieved 2017-05-01.
- ^ a b Ben-Dor, Amir; Shamir, Ron; Yakhini, Zohar (1999-10-01). "Clustering Gene Expression Patterns". Journal of Computational Biology. 6 (3–4): 281–297. CiteSeerX 10.1.1.34.5341. doi:10.1089/106652799318274. ISSN 1066-5277. PMID 10582567.
- ^ Valafar, Faramarz (2002-12-01). "Pattern Recognition Techniques in Microarray Data Analysis". Annals of the New York Academy of Sciences. 980 (1): 41–64. Bibcode:2002NYASA.980...41V. CiteSeerX 10.1.1.199.6445. doi:10.1111/j.1749-6632.2002.tb04888.x. ISSN 1749-6632. PMID 12594081. S2CID 343093.
- ^ Valafar F. Pattern recognition techniques in microarray data analysis. Annals of the New York Academy of Sciences. 2002 Dec 1;980(1):41-64.
- ^ Ahmed, Mohamed N.; Yamany, Sameh M.; Mohamed, Nevin; Farag, Aly A.; Moriarty, Thomas (2002). "A Modified Fuzzy C-Means Algorithm for Bias Field Estimation and Segmentation of MRI Data" (PDF). IEEE Transactions on Medical Imaging. 21 (3): 193–199. CiteSeerX 10.1.1.331.9742. doi:10.1109/42.996338. PMID 11989844. S2CID 8480349. Archived from the original (PDF) on 2011-10-02. Retrieved 2011-10-02..
- ^ Banerjee, Tanvi (2014). "Day or Night Activity Recognition From Video Using Fuzzy Clustering Techniques". IEEE Transactions on Fuzzy Systems. 22 (3): 483–493. CiteSeerX 10.1.1.652.2819. doi:10.1109/TFUZZ.2013.2260756. S2CID 11606344.
- ^ Alireza, Kashani; Kashani, Amir; Milani, Nargess; Akhlaghi, Peyman; Khezri, Kaveh (2008). "Robust Color Classification Using Fuzzy Reasoning and Genetic Algorithms in RoboCup Soccer Leagues". RoboCup 2007: Robot Soccer World Cup XI. Lecture Notes in Computer Science. Vol. 5001. pp. 548–555. doi:10.1007/978-3-540-68847-1_59. ISBN 978-3-540-68846-4.
{{cite book}}:|journal=ignored (help) - ^ "Fuzzy Clustering - MATLAB & Simulink". www.mathworks.com. Retrieved 2017-05-03.
- ^ Lecca, Paola (2011). Systemic Approaches in Bioinformatics and Computational Systems Biology. IGI Global. p. 9. ISBN 9781613504369.







![{\displaystyle W=w_{i,j}\in [0,1],\;i=1,...,n,\;j=1,...,c}](https://wikimedia.org/enwiki/api/rest_v1/media/math/render/svg/4c27fad78fc48f1a8cbf22ee5cbb4c4389665fab)








