Caspase-activated DNase: Difference between revisions
m WP:CHECKWIKI error fix for #61. Punctuation goes before References. Do general fixes if a problem exists. - using AWB |
SimLibrarian (talk | contribs) m Updated short description #article-change-desc Tags: Mobile edit Mobile app edit iOS app edit |
||
| (44 intermediate revisions by 23 users not shown) | |||
| Line 1: | Line 1: | ||
{{Short description|Protein found in humans}} |
|||
{{PBB|geneid=1677}} |
|||
{{cs1 config|name-list-style=vanc|display-authors=6}} |
|||
{{Infobox_gene}} |
|||
{{Infobox protein family |
{{Infobox protein family |
||
| Symbol = DFF40 |
| Symbol = DFF40 |
||
| Line 5: | Line 7: | ||
| image = PDB 1v0d EBI.jpg |
| image = PDB 1v0d EBI.jpg |
||
| width = |
| width = |
||
| caption = |
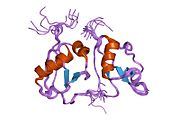
| caption = Crystal structure of caspase-activated DNAse (CAD) |
||
| Pfam = PF09230 |
| Pfam = PF09230 |
||
| Pfam_clan = |
| Pfam_clan = |
||
| Line 20: | Line 22: | ||
}} |
}} |
||
'''Caspase- |
'''Caspase-activated DNase''' ('''CAD''') or '''DNA fragmentation factor subunit beta''' is a [[protein]] that in humans is encoded by the ''DFFB'' [[gene]].<ref name="pmid9108473">{{cite journal | vauthors = Liu X, Zou H, Slaughter C, Wang X | title = DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis | journal = Cell | volume = 89 | issue = 2 | pages = 175–84 | date = April 1997 | pmid = 9108473 | doi = 10.1016/S0092-8674(00)80197-X | s2cid = 14800864 | doi-access = free }}</ref><ref name="pmid9560346">{{cite journal | vauthors = Halenbeck R, MacDonald H, Roulston A, Chen TT, Conroy L, Williams LT | title = CPAN, a human nuclease regulated by the caspase-sensitive inhibitor DFF45 | journal = Current Biology | volume = 8 | issue = 9 | pages = 537–40 | date = April 1998 | pmid = 9560346 | doi = 10.1016/S0960-9822(98)79298-X | s2cid = 9837862 | doi-access = free | bibcode = 1998CBio....8..537H }}</ref><ref name="entrez">{{cite web | title = Entrez Gene: DFFB DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase)| url = https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=1677}}</ref> It breaks up the DNA during apoptosis and promotes cell differentiation. It is usually an inactive monomer inhibited by ICAD. This is cleaved before dimerization. |
||
== Function == |
== Function == |
||
Apoptosis is a cell |
Apoptosis is a cell self-destruct process that removes toxic and/or useless cells during mammalian development and other life processes. The apoptotic process is accompanied by shrinkage and fragmentation of the cells and nuclei and degradation of the chromosomal DNA into nucleosomal units. DNA fragmentation factor (DFF) is a heterodimeric protein of 40-kD (DFFB) and 45-kD ([[DFFA]]) subunits. DFFA is the substrate for caspase-3 and triggers DNA fragmentation during apoptosis. DFF becomes activated when DFFA is cleaved by caspase-3. The cleaved fragments of DFFA dissociate from DFFB, the active component of DFF. DFFB has been found to trigger both DNA fragmentation and chromatin condensation during apoptosis. Multiple alternatively spliced transcript variants encoding distinct isoforms have been found for this gene, but the biological validity of some variants has not been determined.<ref name="entrez" /> |
||
[[Image:CAD and ICAD forms.png|thumb|CAD and ICAD forms]] |
[[Image:CAD and ICAD forms.png|thumb|CAD and ICAD forms]] |
||
Despite this gene being present in every cell, this protein is only expressed in different tissues and cell variety such as pancreas, heart, colon, leukocytes, prostate, ovary, placenta, kidney, spleen and thymus.<ref name=Bio.davidson>{{cite web|last1=Davidson College|url=http://www.bio.davidson.edu/courses/immunology/students/spring2006/ryan/yfip.html |title = Caspase Activated Deoxyribonuclease (CAD) | |
Despite this gene being present in every cell, this protein is only expressed in different tissues and cell variety such as pancreas, heart, colon, leukocytes, prostate, ovary, placenta, kidney, spleen and thymus.<ref name=Bio.davidson>{{cite web|last1=Davidson College|url=http://www.bio.davidson.edu/courses/immunology/students/spring2006/ryan/yfip.html |title = Caspase Activated Deoxyribonuclease (CAD) |access-date=21 Jan 2016}}</ref> |
||
It is also known as |
It is also known as caspase activated nuclease (CPAN), dna fragmentation factor 40 (DFF-40), DFF2 and DFFB. Besides, there are other nomenclatures as a result of combining the previous ones.<ref name=Bio.davidson /><ref name=contribution>{{cite journal | vauthors = Yuste VJ, Sánchez-López I, Solé C, Moubarak RS, Bayascas JR, Dolcet X, Encinas M, Susin SA, Comella JX | title = The contribution of apoptosis-inducing factor, caspase-activated DNase, and inhibitor of caspase-activated DNase to the nuclear phenotype and DNA degradation during apoptosis | journal = The Journal of Biological Chemistry | volume = 280 | issue = 42 | pages = 35670–83 | date = October 2005 | pmid = 16049016 | doi = 10.1074/jbc.M504015200 | doi-access = free }}</ref><ref name=specific>{{cite journal | vauthors = Sakahira H, Iwamatsu A, Nagata S | title = Specific chaperone-like activity of inhibitor of caspase-activated DNase for caspase-activated DNase | journal = The Journal of Biological Chemistry | volume = 275 | issue = 11 | pages = 8091–6 | date = March 2000 | pmid = 10713130 | doi = 10.1074/jbc.275.11.8091 | doi-access = free }}</ref><ref name=functional>{{cite journal | vauthors = Sakahira H, Enari M, Nagata S | title = Functional differences of two forms of the inhibitor of caspase-activated DNase, ICAD-L, and ICAD-S | journal = The Journal of Biological Chemistry | volume = 274 | issue = 22 | pages = 15740–4 | date = May 1999 | pmid = 10336474 | doi = 10.1074/jbc.274.22.15740 | doi-access = free }}</ref> |
||
== Structure == |
== Structure == |
||
This heterodimer is an [[endonuclease]]<ref name=contribution /><ref name=lupus>{{cite journal | vauthors = Jog NR, Frisoni L, Shi Q, Monestier M, Hernandez S, Craft J, Prak ET, Caricchio R | title = Caspase-activated DNase is required for maintenance of tolerance to lupus nuclear autoantigens | journal = Arthritis and Rheumatism | volume = 64 | issue = 4 | pages = 1247–56 | date = April 2012 | pmid = 22127758 | pmc = 3292632 | doi = 10.1002/art.33448 }}</ref><ref name=subunit>{{cite journal | vauthors = Widlak P, Lanuszewska J, Cary RB, Garrard WT | title = Subunit structures and stoichiometries of human DNA fragmentation factor proteins before and after induction of apoptosis | journal = The Journal of Biological Chemistry | volume = 278 | issue = 29 | pages = 26915–22 | date = July 2003 | pmid = 12748178 | doi = 10.1074/jbc.M303807200 }}</ref> with a high content of [[cysteine]] residues.<ref name=functional /> It remains inactive in growing cells while it is associated with its inhibitor (ICAD, DNA fragmentation factor 45 kDa subunit, [[DFFA]] or DFF45) resulting into a complex ICAD-CAD.<ref name=Bio.davidson /><ref name=contribution /><ref name=functional /><ref name=lupus /><ref name=structural>{{cite journal | vauthors = Reh S, Korn C, Gimadutdinow O, Meiss G | title = Structural basis for stable DNA complex formation by the caspase-activated DNase | journal = The Journal of Biological Chemistry | volume = 280 | issue = 50 | pages = 41707–15 | date = December 2005 | pmid = 16236713 | doi = 10.1074/jbc.m509133200 }}</ref><ref name=cleavage>{{cite journal | vauthors = Widlak P, Li P, Wang X, Garrard WT | title = Cleavage preferences of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease) on naked DNA and chromatin substrates | journal = The Journal of Biological Chemistry | volume = 275 | issue = 11 | pages = 8226–32 | date = March 2000 | pmid = 10713148 | doi = 10.1074/jbc.275.11.8226 }}</ref><ref name=direct>{{cite journal | vauthors = Sharif-Askari E, Alam A, Rhéaume E, Beresford PJ, Scotto C, Sharma K, Lee D, DeWolf WE, Nuttall ME, Lieberman J, Sékaly RP | title = Direct cleavage of the human DNA fragmentation factor-45 by granzyme B induces caspase-activated DNase release and DNA fragmentation | journal = The EMBO Journal | volume = 20 | issue = 12 | pages = 3101–13 | date = June 2001 | pmid = 11406587 | pmc = 150191 | doi = 10.1093/emboj/20.12.3101 }}</ref><ref name=activation>{{cite journal | vauthors = Liu X, Zou H, Widlak P, Garrard W, Wang X | title = Activation of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease). Oligomerization and direct interaction with histone H1 | journal = The Journal of Biological Chemistry | volume = 274 | issue = 20 | pages = 13836–40 | date = May 1999 | pmid = 10318789 | doi = 10.1074/jbc.274.20.13836 }}</ref> Their dissociation allows DFF40 to oligomerize to form a large functional complex which is by itself an active DNase.<ref name=functional /><ref name=lupus /><ref name=cleavage /><ref name=direct /><ref name=activation /> |
This heterodimer is an [[endonuclease]]<ref name=contribution /><ref name=lupus>{{cite journal | vauthors = Jog NR, Frisoni L, Shi Q, Monestier M, Hernandez S, Craft J, Prak ET, Caricchio R | title = Caspase-activated DNase is required for maintenance of tolerance to lupus nuclear autoantigens | journal = Arthritis and Rheumatism | volume = 64 | issue = 4 | pages = 1247–56 | date = April 2012 | pmid = 22127758 | pmc = 3292632 | doi = 10.1002/art.33448 }}</ref><ref name=subunit>{{cite journal | vauthors = Widlak P, Lanuszewska J, Cary RB, Garrard WT | title = Subunit structures and stoichiometries of human DNA fragmentation factor proteins before and after induction of apoptosis | journal = The Journal of Biological Chemistry | volume = 278 | issue = 29 | pages = 26915–22 | date = July 2003 | pmid = 12748178 | doi = 10.1074/jbc.M303807200 | doi-access = free }}</ref> with a high content of [[cysteine]] residues.<ref name=functional /> It remains inactive in growing cells while it is associated with its inhibitor (ICAD, DNA fragmentation factor 45 kDa subunit, [[DFFA]] or DFF45) resulting into a complex ICAD-CAD.<ref name=Bio.davidson /><ref name=contribution /><ref name=functional /><ref name=lupus /><ref name=structural>{{cite journal | vauthors = Reh S, Korn C, Gimadutdinow O, Meiss G | title = Structural basis for stable DNA complex formation by the caspase-activated DNase | journal = The Journal of Biological Chemistry | volume = 280 | issue = 50 | pages = 41707–15 | date = December 2005 | pmid = 16236713 | doi = 10.1074/jbc.m509133200 | doi-access = free }}</ref><ref name=cleavage>{{cite journal | vauthors = Widlak P, Li P, Wang X, Garrard WT | title = Cleavage preferences of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease) on naked DNA and chromatin substrates | journal = The Journal of Biological Chemistry | volume = 275 | issue = 11 | pages = 8226–32 | date = March 2000 | pmid = 10713148 | doi = 10.1074/jbc.275.11.8226 | doi-access = free }}</ref><ref name=direct>{{cite journal | vauthors = Sharif-Askari E, Alam A, Rhéaume E, Beresford PJ, Scotto C, Sharma K, Lee D, DeWolf WE, Nuttall ME, Lieberman J, Sékaly RP | title = Direct cleavage of the human DNA fragmentation factor-45 by granzyme B induces caspase-activated DNase release and DNA fragmentation | journal = The EMBO Journal | volume = 20 | issue = 12 | pages = 3101–13 | date = June 2001 | pmid = 11406587 | pmc = 150191 | doi = 10.1093/emboj/20.12.3101 }}</ref><ref name=activation>{{cite journal | vauthors = Liu X, Zou H, Widlak P, Garrard W, Wang X | title = Activation of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease). Oligomerization and direct interaction with histone H1 | journal = The Journal of Biological Chemistry | volume = 274 | issue = 20 | pages = 13836–40 | date = May 1999 | pmid = 10318789 | doi = 10.1074/jbc.274.20.13836 | doi-access = free }}</ref> Their dissociation allows DFF40 to oligomerize to form a large functional complex which is by itself an active DNase.<ref name=functional /><ref name=lupus /><ref name=cleavage /><ref name=direct /><ref name=activation /> |
||
=== DFF40 subunit or CAD === |
=== DFF40 subunit or CAD === |
||
It |
It weighs 40 kDa. Moreover, it contains three domains making up a CAD monomer: C1 or N-terminal CAD; C2 which conform three separate α chains and, at last, C3 which is the largest and functionally the most important. What is more, combining C3’s amino acids leads to 5 α helices, 4 β lamina and a loop at the catalytic C-terminal which interact with each other. Therefore, a cavity (active site) where DNA can fit is produced, even though there is another binding region responsible for stable DNA complex during its fragmentation.<ref name=Bio.davidson /><ref name=structural /><ref name=structure>{{cite journal | vauthors = Uegaki K, Otomo T, Sakahira H, Shimizu M, Yumoto N, Kyogoku Y, Nagata S, Yamazaki T | title = Structure of the CAD domain of caspase-activated DNase and interaction with the CAD domain of its inhibitor | journal = Journal of Molecular Biology | volume = 297 | issue = 5 | pages = 1121–8 | date = April 2000 | pmid = 10764577 | doi = 10.1006/jmbi.2000.3643 }}</ref> |
||
=== DFF45 subunit or ICAD === |
=== DFF45 subunit or ICAD === |
||
[[DFFA]] is encoded by an alternatively encrypted mRNAs originating two distinct forms: short (ICAD-S) and long (ICAD-L), which act like a specific chaperone ensuring the correct |
[[DFFA]] is encoded by an alternatively encrypted mRNAs originating two distinct forms: short (ICAD-S) and long (ICAD-L), which act like a specific chaperone ensuring the correct CAD's folding<ref name=specific /><ref name=functional /><ref name=activation /> Besides, it contains two aspartic acid residues (Asp117 and Asp224) where CAD is identified and, consequently, it stays bounded until [[Caspase-3]] splits this union.<ref name=specific /><ref name=structural /> |
||
== Activation process == |
== Activation process == |
||
Per usual in non-apoptotic growing cells caspase activated dnase is held in check inactivated in the cytoplasm thanks to the association with its inhibitor, inhibitor of caspase-activated DNase (ICAD) also known as DNA fragmentation factor 45 kDa (DFF45). |
Per usual in non-apoptotic growing cells caspase activated dnase is held in check inactivated in the cytoplasm thanks to the association with its inhibitor, inhibitor of caspase-activated DNase (ICAD) also known as DNA fragmentation factor 45 kDa (DFF45). |
||
ICAD is encoded by alternatively spliced mRNAs which generate long (ICAD-L) and short (ICAD-S) forms of ICAD. Therefore, ICAD has a double function; it acts as a CAD inhibitor and also as a [[chaperone (protein)|chaperone]] for CAD synthesis assisting the correct assembly of the protein.<ref> |
ICAD is encoded by alternatively spliced mRNAs which generate long (ICAD-L) and short (ICAD-S) forms of ICAD. Therefore, ICAD has a double function; it acts as a CAD inhibitor and also as a [[chaperone (protein)|chaperone]] for CAD synthesis assisting the correct assembly of the protein.<ref>{{PDB|1V0D}}; {{cite journal | vauthors = Woo EJ, Kim YG, Kim MS, Han WD, Shin S, Robinson H, Park SY, Oh BH | title = Structural mechanism for inactivation and activation of CAD/DFF40 in the apoptotic pathway | journal = Molecular Cell | volume = 14 | issue = 4 | pages = 531–9 | date = May 2004 | pmid = 15149602 | doi = 10.1016/S1097-2765(04)00258-8 | doi-access = free }}</ref> |
||
ICAD has two caspase recognition sites at Asp117 and Asp224. CAD release from ICAD inhibition is achieved by cleavage of ICAD at these Asp residues by the [[caspase-3]].<ref> |
ICAD has two caspase recognition sites at Asp117 and Asp224. CAD release from ICAD inhibition is achieved by cleavage of ICAD at these Asp residues by the [[caspase-3]].<ref>{{Cite web | url=https://www.ncbi.nlm.nih.gov/gene/836 |title = CASP3 caspase 3 [Homo sapiens (human)] - Gene - NCBI}}</ref> |
||
Caspase-3 is activated in the apoptotic cell.<ref name=contribution /> Caspase-3 activation is a cell requirement during early stages of the skeletal myoblast differentiation. Its [[catalytic site]] involves sulfohydryl group of Cys-285 and the imidazole ring of its His-237. The caspase-3 His-237 stabilizes the target Aspartate causing the break of the association of ICAD and CAD leaving the endonuclease CAD active allowing it to degrade chromosomal DNA. |
Caspase-3 is activated in the apoptotic cell.<ref name=contribution /> Caspase-3 activation is a cell requirement during early stages of the skeletal myoblast differentiation. Its [[catalytic site]] involves sulfohydryl group of Cys-285 and the imidazole ring of its His-237. The caspase-3 His-237 stabilizes the target Aspartate causing the break of the association of ICAD and CAD leaving the endonuclease CAD active allowing it to degrade chromosomal DNA. |
||
| Line 54: | Line 56: | ||
== Interactions == |
== Interactions == |
||
DFFB has been shown to [[Protein-protein interaction|interact]] with [[DFFA]].<ref name=pmid17353931>{{cite journal | vauthors = Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, Vasilescu J, Abu-Farha M, Lambert JP, Duewel HS, Stewart II, Kuehl B, Hogue K, Colwill K, Gladwish K, Muskat B, Kinach R, Adams SL, Moran MF, Morin GB, Topaloglou T, Figeys D |
DFFB has been shown to [[Protein-protein interaction|interact]] with [[DFFA]].<ref name=pmid17353931>{{cite journal | vauthors = Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, Vasilescu J, Abu-Farha M, Lambert JP, Duewel HS, Stewart II, Kuehl B, Hogue K, Colwill K, Gladwish K, Muskat B, Kinach R, Adams SL, Moran MF, Morin GB, Topaloglou T, Figeys D | title = Large-scale mapping of human protein-protein interactions by mass spectrometry | journal = Molecular Systems Biology | volume = 3 | issue = 1 | pages = 89 | year = 2007 | pmid = 17353931 | pmc = 1847948 | doi = 10.1038/msb4100134 }}</ref><ref name=pmid10527860>{{cite journal | vauthors = McCarty JS, Toh SY, Li P | title = Study of DFF45 in its role of chaperone and inhibitor: two independent inhibitory domains of DFF40 nuclease activity | journal = Biochemical and Biophysical Research Communications | volume = 264 | issue = 1 | pages = 176–80 | date = October 1999 | pmid = 10527860 | doi = 10.1006/bbrc.1999.1497 }}</ref> |
||
== Cell |
== Cell differentiation == |
||
Caspase 3 is responsible for [[cellular differentiation]], although it is unclear how this kind of protein can promote the cell [[apoptosis]]. Caspase signals resulting from the activation of nuclease CAD indicate that the cell differentiation is due to a CAD modification in chromatin structure. |
Caspase 3 is responsible for [[cellular differentiation]], although it is unclear how this kind of protein can promote the cell [[apoptosis]]. Caspase signals resulting from the activation of nuclease CAD indicate that the cell differentiation is due to a CAD modification in chromatin structure. |
||
CAD leads to the initiation of the DNA strand breakage, which occurs during terminal differentiation of some cell, such as skeletal muscle cell. Targeting of p21 promoter is responsible for inducing cell differentiation, which is promoted by modifying the DNA nuclear microenvironment.<ref>{{cite journal | vauthors = Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA | title = Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 107 | issue = 9 | pages = 4230–5 | date = March 2010 | pmid = 20160104 | pmc = 2840077 | doi = 10.1073/pnas.0913089107 | bibcode = 2010PNAS..107.4230L }}</ref> |
CAD leads to the initiation of the DNA strand breakage, which occurs during terminal differentiation of some cell, such as skeletal muscle cell. Targeting of p21 promoter is responsible for inducing cell differentiation, which is promoted by modifying the DNA nuclear microenvironment.<ref>{{cite journal | vauthors = Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA | title = Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 107 | issue = 9 | pages = 4230–5 | date = March 2010 | pmid = 20160104 | pmc = 2840077 | doi = 10.1073/pnas.0913089107 | bibcode = 2010PNAS..107.4230L | doi-access = free }}</ref> |
||
The cell diversity is originated by cell differentiation, which has been attributed to the activation of specific transcription factors. It also depends on the activity of a protein or a common signal. The factor that seems to induce more cell differentiation is caspase-3 protease.<ref>{{cite journal | vauthors = Fernando P, Megeney LA | title = Is caspase-dependent apoptosis only cell differentiation taken to the extreme? | journal = FASEB Journal | volume = 21 | issue = 1 | pages = 8–17 | date = January 2007 | pmid = 17093139 | doi = 10.1096/fj.06-5912hyp }}</ref> This was identified as the penultimate stage of apoptosis pathways cell. |
The cell diversity is originated by cell differentiation, which has been attributed to the activation of specific transcription factors. It also depends on the activity of a protein or a common signal. The factor that seems to induce more cell differentiation is caspase-3 protease.<ref>{{cite journal | vauthors = Fernando P, Megeney LA | title = Is caspase-dependent apoptosis only cell differentiation taken to the extreme? | journal = FASEB Journal | volume = 21 | issue = 1 | pages = 8–17 | date = January 2007 | pmid = 17093139 | doi = 10.1096/fj.06-5912hyp | doi-access = free | s2cid = 11933880 }}</ref> This was identified as the penultimate stage of apoptosis pathways cell. |
||
Some studies have shown that this differentiation is due to many CAD kinase substrates. Referring to the example of skeletal cells, their differentiation is associated to cleavage of the kinase MST1.<ref>{{cite journal | vauthors = Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA | title = Caspase 3 activity is required for skeletal muscle differentiation | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 99 | issue = 17 | pages = 11025–30 | date = August 2002 | pmid = 12177420 | doi = 10.1073/pnas.162172899 | bibcode = 2002PNAS...9911025F }}</ref> |
Some studies have shown that this differentiation is due to many CAD kinase substrates. Referring to the example of skeletal cells, their differentiation is associated to cleavage of the kinase MST1.<ref>{{cite journal | vauthors = Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA | title = Caspase 3 activity is required for skeletal muscle differentiation | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 99 | issue = 17 | pages = 11025–30 | date = August 2002 | pmid = 12177420 | doi = 10.1073/pnas.162172899 | bibcode = 2002PNAS...9911025F | pmc=123204| doi-access = free }}</ref> |
||
Moreover, it has been seen that CAD participates in the formation of genome whose DNA breaks during early stages of the cell differentiation. Besides, Caspase 3 induces DNA breaks in the promoter of the factor p21 and this strand breakup is related to p21 gene expression. |
Moreover, it has been seen that CAD participates in the formation of genome whose DNA breaks during early stages of the cell differentiation. Besides, Caspase 3 induces DNA breaks in the promoter of the factor p21 and this strand breakup is related to p21 gene expression. |
||
== Cell apoptotic death == |
== Cell apoptotic death == |
||
The protein |
The protein caspase DNase is an endonuclease involved in the cell apoptotic process that facilitates the DNA breakup.<ref>{{cite journal|author-link2=Wong Chi-huey|author-link3=Yuan-Pern Lee | vauthors = Lai SK, Wong CH, Lee YP, Li HY | title = Caspase-3-mediated degradation of condensin Cap-H regulates mitotic cell death | journal = Cell Death and Differentiation | volume = 18 | issue = 6 | pages = 996–1004 | date = June 2011 | pmid = 21151026 | doi = 10.1038/cdd.2010.165 | pmc=3131938}}</ref> Cell apoptotic death is a process executed by [[cysteine]] [[proteases]]<ref>{{cite journal | vauthors = Marsden VS, O'Connor L, O'Reilly LA, Silke J, Metcalf D, Ekert PG, Huang DC, Cecconi F, Kuida K, Tomaselli KJ, Roy S, Nicholson DW, Vaux DL, Bouillet P, Adams JM, Strasser A | title = Apoptosis initiated by Bcl-2-regulated caspase activation independently of the cytochrome c/Apaf-1/caspase-9 apoptosome | journal = Nature | volume = 419 | issue = 6907 | pages = 634–7 | date = October 2002 | pmid = 12374983 | doi = 10.1038/nature01101 | bibcode = 2002Natur.419..634M | s2cid = 4415828 }}</ref> that allows the animals to keep their [[homeostasis]], also regulated by other mechanisms such as the growth and cell differentiation. This biological response is characterized by the chromosomal [[DNA]]’s degradation in tiny fragments within the nucleus of the cell.<ref name="nature.com">{{cite journal | vauthors = Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S | title = A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD | journal = Nature | volume = 391 | issue = 6662 | pages = 43–50 | date = January 1998 | pmid = 9422506 | doi = 10.1038/34112 | bibcode = 1998Natur.391...43E | s2cid = 4407426 }}</ref> After many investigations and research, it was possible to ensure that Caspase-activated DNase is the main responsible of this destruction due to a long list of stimuli. |
||
One of the experiments carried out by the investigators in order to prove that theory was based on the introduction of mutated form of this protein inside both TF-1 human cells and [[Jurkat cells]], which had already reacted to the usual (not mutated) form of the endonuclease and they had dead of apoptosis. As a result, these cells died taking into account this genetic modification but they did not show DNA breakup. This was the key evidence to prove that the CAD form is implicated in this part of the process because without its contribution the fragmentation did not take place.<ref>{{cite journal | vauthors = McIlroy D, Sakahira H, Talanian RV, Nagata S | title = Involvement of caspase 3-activated DNase in internucleosomal DNA cleavage induced by diverse apoptotic stimuli | journal = Oncogene | volume = 18 | issue = 31 | pages = 4401–8 | date = August 1999 | pmid = 10442630 | doi = 10.1038/sj.onc.1202868 }}</ref> |
One of the experiments carried out by the investigators in order to prove that theory was based on the introduction of mutated form of this protein inside both TF-1 human cells and [[Jurkat cells]], which had already reacted to the usual (not mutated) form of the endonuclease and they had dead of apoptosis. As a result, these cells died taking into account this genetic modification but they did not show DNA breakup. This was the key evidence to prove that the CAD form is implicated in this part of the process because without its contribution the fragmentation did not take place.<ref>{{cite journal | vauthors = McIlroy D, Sakahira H, Talanian RV, Nagata S | title = Involvement of caspase 3-activated DNase in internucleosomal DNA cleavage induced by diverse apoptotic stimuli | journal = Oncogene | volume = 18 | issue = 31 | pages = 4401–8 | date = August 1999 | pmid = 10442630 | doi = 10.1038/sj.onc.1202868 | doi-access = free }}</ref> |
||
Later, it was found |
Later, it was found that the way how this protein induces the DNA breakup is explained by its forms CAD and ICAD, which facilitate both the entry and exit in the nucleus of the cell.<ref name="nature.com"/> |
||
== References == |
== References == |
||
{{ |
{{reflist|33em}} |
||
== Further reading == |
== Further reading == |
||
{{refbegin|33em}} |
{{refbegin|33em}} |
||
* {{cite video | year = 2009 | title = Induction of Apoptosis | url = |
* {{cite video | year = 2009 | title = Induction of Apoptosis | url = https://www.youtube.com/watch?v=LCEVqrkPKlA | medium = Video | publisher = [[Garland Science]] / [[YouTube]] }} From {{cite book | last1 = Murphy | first1 = Kenneth | last2 = Travers | first2 = Paul | last3 = Waldport | first3 = Mark | last4 = Ehrenstein | first4 = Michael | title = Laneway's Immunobiology | date = 2008 | publisher = Garland Science | location = New York | isbn = 978-0-8153-4123-9 | edition = 7th | url-access = registration | url = https://archive.org/details/janewaysimmunobi00murp }} |
||
* {{cite journal | vauthors = Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S | title = A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD | journal = Nature | volume = 391 | issue = 6662 | pages = 43–50 | date = January 1998 | pmid = 9422506 | doi = 10.1038/34112 }} |
* {{cite journal | vauthors = Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S | title = A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD | journal = Nature | volume = 391 | issue = 6662 | pages = 43–50 | date = January 1998 | pmid = 9422506 | doi = 10.1038/34112 | bibcode = 1998Natur.391...43E | s2cid = 4407426 }} |
||
* {{cite journal | vauthors = Liu X, Li P, Widlak P, Zou H, Luo X, Garrard WT, Wang X | title = The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 95 | issue = 15 | pages = 8461–6 | date = July 1998 | pmid = 9671700 | pmc = 21098 | doi = 10.1073/pnas.95.15.8461 }} |
* {{cite journal | vauthors = Liu X, Li P, Widlak P, Zou H, Luo X, Garrard WT, Wang X | title = The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 95 | issue = 15 | pages = 8461–6 | date = July 1998 | pmid = 9671700 | pmc = 21098 | doi = 10.1073/pnas.95.15.8461 | bibcode = 1998PNAS...95.8461L | doi-access = free }} |
||
* {{cite journal | vauthors = Mukae N, Enari M, Sakahira H, Fukuda Y, Inazawa J, Toh H, Nagata S | title = Molecular cloning and characterization of human caspase-activated DNase | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 95 | issue = 16 | pages = 9123–8 | date = August 1998 | pmid = 9689044 | pmc = 21302 | doi = 10.1073/pnas.95.16.9123 }} |
* {{cite journal | vauthors = Mukae N, Enari M, Sakahira H, Fukuda Y, Inazawa J, Toh H, Nagata S | title = Molecular cloning and characterization of human caspase-activated DNase | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 95 | issue = 16 | pages = 9123–8 | date = August 1998 | pmid = 9689044 | pmc = 21302 | doi = 10.1073/pnas.95.16.9123 | bibcode = 1998PNAS...95.9123M | doi-access = free }} |
||
* {{cite journal | vauthors = Gu J, Dong RP, Zhang C, McLaughlin DF, Wu MX, Schlossman SF | title = Functional interaction of DFF35 and DFF45 with caspase-activated DNA fragmentation nuclease DFF40 | journal = The Journal of Biological Chemistry | volume = 274 | issue = 30 | pages = 20759–62 | date = July 1999 | pmid = 10409614 | doi = 10.1074/jbc.274.30.20759 }} |
* {{cite journal | vauthors = Gu J, Dong RP, Zhang C, McLaughlin DF, Wu MX, Schlossman SF | title = Functional interaction of DFF35 and DFF45 with caspase-activated DNA fragmentation nuclease DFF40 | journal = The Journal of Biological Chemistry | volume = 274 | issue = 30 | pages = 20759–62 | date = July 1999 | pmid = 10409614 | doi = 10.1074/jbc.274.30.20759 | doi-access = free }} |
||
* {{cite journal | vauthors = McCarty JS, Toh SY, Li P | title = Study of DFF45 in its role of chaperone and inhibitor: two independent inhibitory domains of DFF40 nuclease activity | journal = Biochemical and Biophysical Research Communications | volume = 264 | issue = 1 | pages = 176–80 | date = October 1999 | pmid = 10527860 | doi = 10.1006/bbrc.1999.1497 }} |
* {{cite journal | vauthors = McCarty JS, Toh SY, Li P | title = Study of DFF45 in its role of chaperone and inhibitor: two independent inhibitory domains of DFF40 nuclease activity | journal = Biochemical and Biophysical Research Communications | volume = 264 | issue = 1 | pages = 176–80 | date = October 1999 | pmid = 10527860 | doi = 10.1006/bbrc.1999.1497 }} |
||
* {{cite journal | vauthors = McCarty JS, Toh SY, Li P | title = Multiple domains of DFF45 bind synergistically to DFF40: roles of caspase cleavage and sequestration of activator domain of DFF40 | journal = Biochemical and Biophysical Research Communications | volume = 264 | issue = 1 | pages = 181–5 | date = October 1999 | pmid = 10527861 | doi = 10.1006/bbrc.1999.1498 }} |
* {{cite journal | vauthors = McCarty JS, Toh SY, Li P | title = Multiple domains of DFF45 bind synergistically to DFF40: roles of caspase cleavage and sequestration of activator domain of DFF40 | journal = Biochemical and Biophysical Research Communications | volume = 264 | issue = 1 | pages = 181–5 | date = October 1999 | pmid = 10527861 | doi = 10.1006/bbrc.1999.1498 }} |
||
* {{cite journal | vauthors = Lugovskoy AA, Zhou P, Chou JJ, McCarty JS, Li P, Wagner G | title = Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis | journal = Cell | volume = 99 | issue = 7 | pages = 747–55 | date = December 1999 | pmid = 10619428 | doi = 10.1016/S0092-8674(00)81672-4 }} |
* {{cite journal | vauthors = Lugovskoy AA, Zhou P, Chou JJ, McCarty JS, Li P, Wagner G | title = Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis | journal = Cell | volume = 99 | issue = 7 | pages = 747–55 | date = December 1999 | pmid = 10619428 | doi = 10.1016/S0092-8674(00)81672-4 | doi-access = free }} |
||
* {{cite journal | vauthors = Judson H, van Roy N, Strain L, Vandesompele J, Van Gele M, Speleman F, Bonthron DT | title = Structure and mutation analysis of the gene encoding DNA fragmentation factor 40 (caspase-activated nuclease), a candidate neuroblastoma tumour suppressor gene | journal = Human Genetics | volume = 106 | issue = 4 | pages = 406–13 | date = April 2000 | pmid = 10830907 | doi = 10.1007/s004390000257 }} |
* {{cite journal | vauthors = Judson H, van Roy N, Strain L, Vandesompele J, Van Gele M, Speleman F, Bonthron DT | title = Structure and mutation analysis of the gene encoding DNA fragmentation factor 40 (caspase-activated nuclease), a candidate neuroblastoma tumour suppressor gene | journal = Human Genetics | volume = 106 | issue = 4 | pages = 406–13 | date = April 2000 | pmid = 10830907 | doi = 10.1007/s004390000257 | s2cid = 38271068 }} |
||
* {{cite journal | vauthors = Otomo T, Sakahira H, Uegaki K, Nagata S, Yamazaki T | title = Structure of the heterodimeric complex between CAD domains of CAD and ICAD | journal = Nature Structural Biology | volume = 7 | issue = 8 | pages = 658–62 | date = August 2000 | pmid = 10932250 | doi = 10.1038/77957 }} |
* {{cite journal | vauthors = Otomo T, Sakahira H, Uegaki K, Nagata S, Yamazaki T | title = Structure of the heterodimeric complex between CAD domains of CAD and ICAD | journal = Nature Structural Biology | volume = 7 | issue = 8 | pages = 658–62 | date = August 2000 | pmid = 10932250 | doi = 10.1038/77957 | s2cid = 12925074 }} |
||
* {{cite journal | vauthors = Durrieu F, Samejima K, Fortune JM, Kandels-Lewis S, Osheroff N, Earnshaw WC | title = DNA topoisomerase IIalpha interacts with CAD nuclease and is involved in chromatin condensation during apoptotic execution | journal = Current Biology | volume = 10 | issue = 15 | pages = 923–6 | year = 2001 | pmid = 10959840 | doi = 10.1016/S0960-9822(00)00620-5 }} |
* {{cite journal | vauthors = Durrieu F, Samejima K, Fortune JM, Kandels-Lewis S, Osheroff N, Earnshaw WC | title = DNA topoisomerase IIalpha interacts with CAD nuclease and is involved in chromatin condensation during apoptotic execution | journal = Current Biology | volume = 10 | issue = 15 | pages = 923–6 | year = 2001 | pmid = 10959840 | doi = 10.1016/S0960-9822(00)00620-5 | s2cid = 17443069 | doi-access = free }} |
||
* {{cite journal | vauthors = Zhou P, Lugovskoy AA, McCarty JS, Li P, Wagner G | title = Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45 | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 98 | issue = 11 | pages = 6051–5 | date = May 2001 | pmid = 11371636 | pmc = 33420 | doi = 10.1073/pnas.111145098 }} |
* {{cite journal | vauthors = Zhou P, Lugovskoy AA, McCarty JS, Li P, Wagner G | title = Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45 | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 98 | issue = 11 | pages = 6051–5 | date = May 2001 | pmid = 11371636 | pmc = 33420 | doi = 10.1073/pnas.111145098 | bibcode = 2001PNAS...98.6051Z | doi-access = free }} |
||
* {{cite journal | vauthors = Nie Z, Phenix BN, Lum JJ, Alam A, Lynch DH, Beckett B, Krammer PH, Sekaly RP, Badley AD | title = HIV-1 protease processes procaspase 8 to cause mitochondrial release of cytochrome c, caspase cleavage and nuclear fragmentation | journal = Cell Death and Differentiation | volume = 9 | issue = 11 | pages = 1172–84 | date = November 2002 | pmid = 12404116 | doi = 10.1038/sj.cdd.4401094 }} |
* {{cite journal | vauthors = Nie Z, Phenix BN, Lum JJ, Alam A, Lynch DH, Beckett B, Krammer PH, Sekaly RP, Badley AD | title = HIV-1 protease processes procaspase 8 to cause mitochondrial release of cytochrome c, caspase cleavage and nuclear fragmentation | journal = Cell Death and Differentiation | volume = 9 | issue = 11 | pages = 1172–84 | date = November 2002 | pmid = 12404116 | doi = 10.1038/sj.cdd.4401094 | s2cid = 38809690 }} |
||
* {{cite journal | vauthors = Hsieh SY, Liaw SF, Lee SN, Hsieh PS, Lin KH, Chu CM, Liaw YF | title = Aberrant caspase-activated DNase (CAD) transcripts in human hepatoma cells | journal = British Journal of Cancer | volume = 88 | issue = 2 | pages = 210–6 | date = January 2003 | pmid = 12610505 | pmc = 2377037 | doi = 10.1038/sj.bjc.6600695 }} |
* {{cite journal | vauthors = Hsieh SY, Liaw SF, Lee SN, Hsieh PS, Lin KH, Chu CM, Liaw YF|author-link7=Yun-Fan Liaw | title = Aberrant caspase-activated DNase (CAD) transcripts in human hepatoma cells | journal = British Journal of Cancer | volume = 88 | issue = 2 | pages = 210–6 | date = January 2003 | pmid = 12610505 | pmc = 2377037 | doi = 10.1038/sj.bjc.6600695 }} |
||
* {{cite journal | vauthors = Liu QL, Kishi H, Ohtsuka K, Muraguchi A | title = Heat shock protein 70 binds caspase-activated DNase and enhances its activity in TCR-stimulated T cells | journal = Blood | volume = 102 | issue = 5 | pages = 1788–96 | date = September 2003 | pmid = 12738667 | doi = 10.1182/blood-2002-11-3499 }} |
* {{cite journal | vauthors = Liu QL, Kishi H, Ohtsuka K, Muraguchi A | title = Heat shock protein 70 binds caspase-activated DNase and enhances its activity in TCR-stimulated T cells | journal = Blood | volume = 102 | issue = 5 | pages = 1788–96 | date = September 2003 | pmid = 12738667 | doi = 10.1182/blood-2002-11-3499 | doi-access = free }} |
||
* {{cite journal | vauthors = Widlak P, Lanuszewska J, Cary RB, Garrard WT | title = Subunit structures and stoichiometries of human DNA fragmentation factor proteins before and after induction of apoptosis | journal = The Journal of Biological Chemistry | volume = 278 | issue = 29 | pages = 26915–22 | date = July 2003 | pmid = 12748178 | doi = 10.1074/jbc.M303807200 }} |
* {{cite journal | vauthors = Widlak P, Lanuszewska J, Cary RB, Garrard WT | title = Subunit structures and stoichiometries of human DNA fragmentation factor proteins before and after induction of apoptosis | journal = The Journal of Biological Chemistry | volume = 278 | issue = 29 | pages = 26915–22 | date = July 2003 | pmid = 12748178 | doi = 10.1074/jbc.M303807200 | doi-access = free }} |
||
* {{cite journal | vauthors = Hillman RT, Green RE, Brenner SE | title = An unappreciated role for RNA surveillance | journal = Genome Biology | volume = 5 | issue = 2 | pages = R8 | year = 2005 | pmid = 14759258 | pmc = 395752 | doi = 10.1186/gb-2004-5-2-r8 }} |
* {{cite journal | vauthors = Hillman RT, Green RE, Brenner SE | title = An unappreciated role for RNA surveillance | journal = Genome Biology | volume = 5 | issue = 2 | pages = R8 | year = 2005 | pmid = 14759258 | pmc = 395752 | doi = 10.1186/gb-2004-5-2-r8 | doi-access = free }} |
||
* {{cite journal | vauthors = Bayascas JR, Yuste VJ, Solé C, Sánchez-López I, Segura MF, Perera R, Comella JX | title = Characterization of splice variants of human caspase-activated DNase with CIDE-N structure and function | journal = FEBS Letters | volume = 566 | issue = |
* {{cite journal | vauthors = Bayascas JR, Yuste VJ, Solé C, Sánchez-López I, Segura MF, Perera R, Comella JX | title = Characterization of splice variants of human caspase-activated DNase with CIDE-N structure and function | journal = FEBS Letters | volume = 566 | issue = 1–3 | pages = 234–40 | date = May 2004 | pmid = 15147901 | doi = 10.1016/j.febslet.2004.04.050 | s2cid = 22464440 | doi-access = free | bibcode = 2004FEBSL.566..234B }} |
||
{{refend}} |
{{refend}} |
||
| Line 103: | Line 105: | ||
{{Esterases}} |
{{Esterases}} |
||
{{Enzymes}} |
{{Enzymes}} |
||
{{Portal bar| |
{{Portal bar|Biology|border=no}} |
||
[[Category:Apoptosis]] |
[[Category:Apoptosis]] |
||
Latest revision as of 19:09, 24 June 2024
| DFFB | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | DFFB, DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase), CAD, CPAN, DFF-40, DFF2, DFF40, DNA fragmentation factor subunit beta | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 601883; MGI: 1196287; HomoloGene: 3241; GeneCards: DFFB; OMA:DFFB - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| DNA fragmentation factor 40 kDa | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystal structure of caspase-activated DNAse (CAD) | |||||||||
| Identifiers | |||||||||
| Symbol | DFF40 | ||||||||
| Pfam | PF09230 | ||||||||
| InterPro | IPR015311 | ||||||||
| SCOP2 | 1v0d / SCOPe / SUPFAM | ||||||||
| |||||||||
Caspase-activated DNase (CAD) or DNA fragmentation factor subunit beta is a protein that in humans is encoded by the DFFB gene.[5][6][7] It breaks up the DNA during apoptosis and promotes cell differentiation. It is usually an inactive monomer inhibited by ICAD. This is cleaved before dimerization.
Function
[edit]Apoptosis is a cell self-destruct process that removes toxic and/or useless cells during mammalian development and other life processes. The apoptotic process is accompanied by shrinkage and fragmentation of the cells and nuclei and degradation of the chromosomal DNA into nucleosomal units. DNA fragmentation factor (DFF) is a heterodimeric protein of 40-kD (DFFB) and 45-kD (DFFA) subunits. DFFA is the substrate for caspase-3 and triggers DNA fragmentation during apoptosis. DFF becomes activated when DFFA is cleaved by caspase-3. The cleaved fragments of DFFA dissociate from DFFB, the active component of DFF. DFFB has been found to trigger both DNA fragmentation and chromatin condensation during apoptosis. Multiple alternatively spliced transcript variants encoding distinct isoforms have been found for this gene, but the biological validity of some variants has not been determined.[7]

Despite this gene being present in every cell, this protein is only expressed in different tissues and cell variety such as pancreas, heart, colon, leukocytes, prostate, ovary, placenta, kidney, spleen and thymus.[8]
It is also known as caspase activated nuclease (CPAN), dna fragmentation factor 40 (DFF-40), DFF2 and DFFB. Besides, there are other nomenclatures as a result of combining the previous ones.[8][9][10][11]
Structure
[edit]This heterodimer is an endonuclease[9][12][13] with a high content of cysteine residues.[11] It remains inactive in growing cells while it is associated with its inhibitor (ICAD, DNA fragmentation factor 45 kDa subunit, DFFA or DFF45) resulting into a complex ICAD-CAD.[8][9][11][12][14][15][16][17] Their dissociation allows DFF40 to oligomerize to form a large functional complex which is by itself an active DNase.[11][12][15][16][17]
DFF40 subunit or CAD
[edit]It weighs 40 kDa. Moreover, it contains three domains making up a CAD monomer: C1 or N-terminal CAD; C2 which conform three separate α chains and, at last, C3 which is the largest and functionally the most important. What is more, combining C3’s amino acids leads to 5 α helices, 4 β lamina and a loop at the catalytic C-terminal which interact with each other. Therefore, a cavity (active site) where DNA can fit is produced, even though there is another binding region responsible for stable DNA complex during its fragmentation.[8][14][18]
DFF45 subunit or ICAD
[edit]DFFA is encoded by an alternatively encrypted mRNAs originating two distinct forms: short (ICAD-S) and long (ICAD-L), which act like a specific chaperone ensuring the correct CAD's folding[10][11][17] Besides, it contains two aspartic acid residues (Asp117 and Asp224) where CAD is identified and, consequently, it stays bounded until Caspase-3 splits this union.[10][14]
Activation process
[edit]Per usual in non-apoptotic growing cells caspase activated dnase is held in check inactivated in the cytoplasm thanks to the association with its inhibitor, inhibitor of caspase-activated DNase (ICAD) also known as DNA fragmentation factor 45 kDa (DFF45).
ICAD is encoded by alternatively spliced mRNAs which generate long (ICAD-L) and short (ICAD-S) forms of ICAD. Therefore, ICAD has a double function; it acts as a CAD inhibitor and also as a chaperone for CAD synthesis assisting the correct assembly of the protein.[19]
ICAD has two caspase recognition sites at Asp117 and Asp224. CAD release from ICAD inhibition is achieved by cleavage of ICAD at these Asp residues by the caspase-3.[20]
Caspase-3 is activated in the apoptotic cell.[9] Caspase-3 activation is a cell requirement during early stages of the skeletal myoblast differentiation. Its catalytic site involves sulfohydryl group of Cys-285 and the imidazole ring of its His-237. The caspase-3 His-237 stabilizes the target Aspartate causing the break of the association of ICAD and CAD leaving the endonuclease CAD active allowing it to degrade chromosomal DNA.
Once the inhibitor is released and in order to properly function, two CAD monomers need to come together to form a functional dimer that has vertical symmetry.
Interactions
[edit]DFFB has been shown to interact with DFFA.[21][22]
Cell differentiation
[edit]Caspase 3 is responsible for cellular differentiation, although it is unclear how this kind of protein can promote the cell apoptosis. Caspase signals resulting from the activation of nuclease CAD indicate that the cell differentiation is due to a CAD modification in chromatin structure.
CAD leads to the initiation of the DNA strand breakage, which occurs during terminal differentiation of some cell, such as skeletal muscle cell. Targeting of p21 promoter is responsible for inducing cell differentiation, which is promoted by modifying the DNA nuclear microenvironment.[23]
The cell diversity is originated by cell differentiation, which has been attributed to the activation of specific transcription factors. It also depends on the activity of a protein or a common signal. The factor that seems to induce more cell differentiation is caspase-3 protease.[24] This was identified as the penultimate stage of apoptosis pathways cell.
Some studies have shown that this differentiation is due to many CAD kinase substrates. Referring to the example of skeletal cells, their differentiation is associated to cleavage of the kinase MST1.[25]
Moreover, it has been seen that CAD participates in the formation of genome whose DNA breaks during early stages of the cell differentiation. Besides, Caspase 3 induces DNA breaks in the promoter of the factor p21 and this strand breakup is related to p21 gene expression.
Cell apoptotic death
[edit]The protein caspase DNase is an endonuclease involved in the cell apoptotic process that facilitates the DNA breakup.[26] Cell apoptotic death is a process executed by cysteine proteases[27] that allows the animals to keep their homeostasis, also regulated by other mechanisms such as the growth and cell differentiation. This biological response is characterized by the chromosomal DNA’s degradation in tiny fragments within the nucleus of the cell.[28] After many investigations and research, it was possible to ensure that Caspase-activated DNase is the main responsible of this destruction due to a long list of stimuli.
One of the experiments carried out by the investigators in order to prove that theory was based on the introduction of mutated form of this protein inside both TF-1 human cells and Jurkat cells, which had already reacted to the usual (not mutated) form of the endonuclease and they had dead of apoptosis. As a result, these cells died taking into account this genetic modification but they did not show DNA breakup. This was the key evidence to prove that the CAD form is implicated in this part of the process because without its contribution the fragmentation did not take place.[29]
Later, it was found that the way how this protein induces the DNA breakup is explained by its forms CAD and ICAD, which facilitate both the entry and exit in the nucleus of the cell.[28]
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000169598 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000029027 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Liu X, Zou H, Slaughter C, Wang X (April 1997). "DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis". Cell. 89 (2): 175–84. doi:10.1016/S0092-8674(00)80197-X. PMID 9108473. S2CID 14800864.
- ^ Halenbeck R, MacDonald H, Roulston A, Chen TT, Conroy L, Williams LT (April 1998). "CPAN, a human nuclease regulated by the caspase-sensitive inhibitor DFF45". Current Biology. 8 (9): 537–40. Bibcode:1998CBio....8..537H. doi:10.1016/S0960-9822(98)79298-X. PMID 9560346. S2CID 9837862.
- ^ a b "Entrez Gene: DFFB DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase)".
- ^ a b c d Davidson College. "Caspase Activated Deoxyribonuclease (CAD)". Retrieved 21 Jan 2016.
- ^ a b c d Yuste VJ, Sánchez-López I, Solé C, Moubarak RS, Bayascas JR, Dolcet X, et al. (October 2005). "The contribution of apoptosis-inducing factor, caspase-activated DNase, and inhibitor of caspase-activated DNase to the nuclear phenotype and DNA degradation during apoptosis". The Journal of Biological Chemistry. 280 (42): 35670–83. doi:10.1074/jbc.M504015200. PMID 16049016.
- ^ a b c Sakahira H, Iwamatsu A, Nagata S (March 2000). "Specific chaperone-like activity of inhibitor of caspase-activated DNase for caspase-activated DNase". The Journal of Biological Chemistry. 275 (11): 8091–6. doi:10.1074/jbc.275.11.8091. PMID 10713130.
- ^ a b c d e Sakahira H, Enari M, Nagata S (May 1999). "Functional differences of two forms of the inhibitor of caspase-activated DNase, ICAD-L, and ICAD-S". The Journal of Biological Chemistry. 274 (22): 15740–4. doi:10.1074/jbc.274.22.15740. PMID 10336474.
- ^ a b c Jog NR, Frisoni L, Shi Q, Monestier M, Hernandez S, Craft J, et al. (April 2012). "Caspase-activated DNase is required for maintenance of tolerance to lupus nuclear autoantigens". Arthritis and Rheumatism. 64 (4): 1247–56. doi:10.1002/art.33448. PMC 3292632. PMID 22127758.
- ^ Widlak P, Lanuszewska J, Cary RB, Garrard WT (July 2003). "Subunit structures and stoichiometries of human DNA fragmentation factor proteins before and after induction of apoptosis". The Journal of Biological Chemistry. 278 (29): 26915–22. doi:10.1074/jbc.M303807200. PMID 12748178.
- ^ a b c Reh S, Korn C, Gimadutdinow O, Meiss G (December 2005). "Structural basis for stable DNA complex formation by the caspase-activated DNase". The Journal of Biological Chemistry. 280 (50): 41707–15. doi:10.1074/jbc.m509133200. PMID 16236713.
- ^ a b Widlak P, Li P, Wang X, Garrard WT (March 2000). "Cleavage preferences of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease) on naked DNA and chromatin substrates". The Journal of Biological Chemistry. 275 (11): 8226–32. doi:10.1074/jbc.275.11.8226. PMID 10713148.
- ^ a b Sharif-Askari E, Alam A, Rhéaume E, Beresford PJ, Scotto C, Sharma K, et al. (June 2001). "Direct cleavage of the human DNA fragmentation factor-45 by granzyme B induces caspase-activated DNase release and DNA fragmentation". The EMBO Journal. 20 (12): 3101–13. doi:10.1093/emboj/20.12.3101. PMC 150191. PMID 11406587.
- ^ a b c Liu X, Zou H, Widlak P, Garrard W, Wang X (May 1999). "Activation of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease). Oligomerization and direct interaction with histone H1". The Journal of Biological Chemistry. 274 (20): 13836–40. doi:10.1074/jbc.274.20.13836. PMID 10318789.
- ^ Uegaki K, Otomo T, Sakahira H, Shimizu M, Yumoto N, Kyogoku Y, et al. (April 2000). "Structure of the CAD domain of caspase-activated DNase and interaction with the CAD domain of its inhibitor". Journal of Molecular Biology. 297 (5): 1121–8. doi:10.1006/jmbi.2000.3643. PMID 10764577.
- ^ PDB: 1V0D; Woo EJ, Kim YG, Kim MS, Han WD, Shin S, Robinson H, et al. (May 2004). "Structural mechanism for inactivation and activation of CAD/DFF40 in the apoptotic pathway". Molecular Cell. 14 (4): 531–9. doi:10.1016/S1097-2765(04)00258-8. PMID 15149602.
- ^ "CASP3 caspase 3 [Homo sapiens (human)] - Gene - NCBI".
- ^ Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, et al. (2007). "Large-scale mapping of human protein-protein interactions by mass spectrometry". Molecular Systems Biology. 3 (1): 89. doi:10.1038/msb4100134. PMC 1847948. PMID 17353931.
- ^ McCarty JS, Toh SY, Li P (October 1999). "Study of DFF45 in its role of chaperone and inhibitor: two independent inhibitory domains of DFF40 nuclease activity". Biochemical and Biophysical Research Communications. 264 (1): 176–80. doi:10.1006/bbrc.1999.1497. PMID 10527860.
- ^ Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA (March 2010). "Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks". Proceedings of the National Academy of Sciences of the United States of America. 107 (9): 4230–5. Bibcode:2010PNAS..107.4230L. doi:10.1073/pnas.0913089107. PMC 2840077. PMID 20160104.
- ^ Fernando P, Megeney LA (January 2007). "Is caspase-dependent apoptosis only cell differentiation taken to the extreme?". FASEB Journal. 21 (1): 8–17. doi:10.1096/fj.06-5912hyp. PMID 17093139. S2CID 11933880.
- ^ Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA (August 2002). "Caspase 3 activity is required for skeletal muscle differentiation". Proceedings of the National Academy of Sciences of the United States of America. 99 (17): 11025–30. Bibcode:2002PNAS...9911025F. doi:10.1073/pnas.162172899. PMC 123204. PMID 12177420.
- ^ Lai SK, Wong CH, Lee YP, Li HY (June 2011). "Caspase-3-mediated degradation of condensin Cap-H regulates mitotic cell death". Cell Death and Differentiation. 18 (6): 996–1004. doi:10.1038/cdd.2010.165. PMC 3131938. PMID 21151026.
- ^ Marsden VS, O'Connor L, O'Reilly LA, Silke J, Metcalf D, Ekert PG, et al. (October 2002). "Apoptosis initiated by Bcl-2-regulated caspase activation independently of the cytochrome c/Apaf-1/caspase-9 apoptosome". Nature. 419 (6907): 634–7. Bibcode:2002Natur.419..634M. doi:10.1038/nature01101. PMID 12374983. S2CID 4415828.
- ^ a b Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S (January 1998). "A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD". Nature. 391 (6662): 43–50. Bibcode:1998Natur.391...43E. doi:10.1038/34112. PMID 9422506. S2CID 4407426.
- ^ McIlroy D, Sakahira H, Talanian RV, Nagata S (August 1999). "Involvement of caspase 3-activated DNase in internucleosomal DNA cleavage induced by diverse apoptotic stimuli". Oncogene. 18 (31): 4401–8. doi:10.1038/sj.onc.1202868. PMID 10442630.
Further reading
[edit]- Induction of Apoptosis (Video). Garland Science / YouTube. 2009. From Murphy K, Travers P, Waldport M, Ehrenstein M (2008). Laneway's Immunobiology (7th ed.). New York: Garland Science. ISBN 978-0-8153-4123-9.
- Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S (January 1998). "A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD". Nature. 391 (6662): 43–50. Bibcode:1998Natur.391...43E. doi:10.1038/34112. PMID 9422506. S2CID 4407426.
- Liu X, Li P, Widlak P, Zou H, Luo X, Garrard WT, et al. (July 1998). "The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis". Proceedings of the National Academy of Sciences of the United States of America. 95 (15): 8461–6. Bibcode:1998PNAS...95.8461L. doi:10.1073/pnas.95.15.8461. PMC 21098. PMID 9671700.
- Mukae N, Enari M, Sakahira H, Fukuda Y, Inazawa J, Toh H, et al. (August 1998). "Molecular cloning and characterization of human caspase-activated DNase". Proceedings of the National Academy of Sciences of the United States of America. 95 (16): 9123–8. Bibcode:1998PNAS...95.9123M. doi:10.1073/pnas.95.16.9123. PMC 21302. PMID 9689044.
- Gu J, Dong RP, Zhang C, McLaughlin DF, Wu MX, Schlossman SF (July 1999). "Functional interaction of DFF35 and DFF45 with caspase-activated DNA fragmentation nuclease DFF40". The Journal of Biological Chemistry. 274 (30): 20759–62. doi:10.1074/jbc.274.30.20759. PMID 10409614.
- McCarty JS, Toh SY, Li P (October 1999). "Study of DFF45 in its role of chaperone and inhibitor: two independent inhibitory domains of DFF40 nuclease activity". Biochemical and Biophysical Research Communications. 264 (1): 176–80. doi:10.1006/bbrc.1999.1497. PMID 10527860.
- McCarty JS, Toh SY, Li P (October 1999). "Multiple domains of DFF45 bind synergistically to DFF40: roles of caspase cleavage and sequestration of activator domain of DFF40". Biochemical and Biophysical Research Communications. 264 (1): 181–5. doi:10.1006/bbrc.1999.1498. PMID 10527861.
- Lugovskoy AA, Zhou P, Chou JJ, McCarty JS, Li P, Wagner G (December 1999). "Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis". Cell. 99 (7): 747–55. doi:10.1016/S0092-8674(00)81672-4. PMID 10619428.
- Judson H, van Roy N, Strain L, Vandesompele J, Van Gele M, Speleman F, et al. (April 2000). "Structure and mutation analysis of the gene encoding DNA fragmentation factor 40 (caspase-activated nuclease), a candidate neuroblastoma tumour suppressor gene". Human Genetics. 106 (4): 406–13. doi:10.1007/s004390000257. PMID 10830907. S2CID 38271068.
- Otomo T, Sakahira H, Uegaki K, Nagata S, Yamazaki T (August 2000). "Structure of the heterodimeric complex between CAD domains of CAD and ICAD". Nature Structural Biology. 7 (8): 658–62. doi:10.1038/77957. PMID 10932250. S2CID 12925074.
- Durrieu F, Samejima K, Fortune JM, Kandels-Lewis S, Osheroff N, Earnshaw WC (2001). "DNA topoisomerase IIalpha interacts with CAD nuclease and is involved in chromatin condensation during apoptotic execution". Current Biology. 10 (15): 923–6. doi:10.1016/S0960-9822(00)00620-5. PMID 10959840. S2CID 17443069.
- Zhou P, Lugovskoy AA, McCarty JS, Li P, Wagner G (May 2001). "Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45". Proceedings of the National Academy of Sciences of the United States of America. 98 (11): 6051–5. Bibcode:2001PNAS...98.6051Z. doi:10.1073/pnas.111145098. PMC 33420. PMID 11371636.
- Nie Z, Phenix BN, Lum JJ, Alam A, Lynch DH, Beckett B, et al. (November 2002). "HIV-1 protease processes procaspase 8 to cause mitochondrial release of cytochrome c, caspase cleavage and nuclear fragmentation". Cell Death and Differentiation. 9 (11): 1172–84. doi:10.1038/sj.cdd.4401094. PMID 12404116. S2CID 38809690.
- Hsieh SY, Liaw SF, Lee SN, Hsieh PS, Lin KH, Chu CM, et al. (January 2003). "Aberrant caspase-activated DNase (CAD) transcripts in human hepatoma cells". British Journal of Cancer. 88 (2): 210–6. doi:10.1038/sj.bjc.6600695. PMC 2377037. PMID 12610505.
- Liu QL, Kishi H, Ohtsuka K, Muraguchi A (September 2003). "Heat shock protein 70 binds caspase-activated DNase and enhances its activity in TCR-stimulated T cells". Blood. 102 (5): 1788–96. doi:10.1182/blood-2002-11-3499. PMID 12738667.
- Widlak P, Lanuszewska J, Cary RB, Garrard WT (July 2003). "Subunit structures and stoichiometries of human DNA fragmentation factor proteins before and after induction of apoptosis". The Journal of Biological Chemistry. 278 (29): 26915–22. doi:10.1074/jbc.M303807200. PMID 12748178.
- Hillman RT, Green RE, Brenner SE (2005). "An unappreciated role for RNA surveillance". Genome Biology. 5 (2): R8. doi:10.1186/gb-2004-5-2-r8. PMC 395752. PMID 14759258.
- Bayascas JR, Yuste VJ, Solé C, Sánchez-López I, Segura MF, Perera R, et al. (May 2004). "Characterization of splice variants of human caspase-activated DNase with CIDE-N structure and function". FEBS Letters. 566 (1–3): 234–40. Bibcode:2004FEBSL.566..234B. doi:10.1016/j.febslet.2004.04.050. PMID 15147901. S2CID 22464440.