Fusobacteriales-1 RNA motif: Difference between revisions
Appearance
Content deleted Content added
Antonipetrov (talk | contribs) m Add Rfam infobox |
m no longer an orphan |
||
| (7 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
{{Short description|Conserved RNA structure}} |
|||
{{primary sources|date=November 2021}} |
|||
{{Infobox rfam |
{{Infobox rfam |
||
| Name = Fusobacteriales-1 |
| Name = Fusobacteriales-1 |
||
| Line 9: | Line 12: | ||
| miRBase = |
| miRBase = |
||
| miRBase_family = |
| miRBase_family = |
||
| RNA_type = [[Cis-regulatory element |
| RNA_type = [[Cis-regulatory element|Cis-reg]] |
||
| Tax_domain = |
| Tax_domain = |
||
| GO = |
| GO = |
||
| Line 24: | Line 27: | ||
}} |
}} |
||
The '''Fusobacteriales-1 RNA motif''' is a conserved [[RNA]] structure that was [[Bioinformatics discovery of non-coding RNAs|discovered by bioinformatics]].<ref name="Weinberg2017b">{{cite journal |vauthors=Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR |title=Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions |journal=Nucleic Acids Res. |volume=45 |issue=18 |pages= |
The '''Fusobacteriales-1 RNA motif''' is a conserved [[RNA]] structure that was [[Bioinformatics discovery of non-coding RNAs|discovered by bioinformatics]].<ref name="Weinberg2017b">{{cite journal |vauthors=Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR |title=Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions |journal=Nucleic Acids Res. |volume=45 |issue=18 |pages=10811–10823 |date=October 2017 |pmid=28977401 |pmc=5737381 |doi=10.1093/nar/gkx699 }}</ref> |
||
Fusobacteriales-1 motif RNAs are found in [[ |
Fusobacteriales-1 motif RNAs are found in [[Fusobacteriales]]. |
||
Most Fusobacteriales-1 RNAs are located upstream of [[gene]]s homologous to the locus FSAG_00736, which encodes a hypothetical protein in ''Fusobacterium peridonticum''. |
Most Fusobacteriales-1 RNAs are located upstream of [[gene]]s homologous to the locus FSAG_00736, which encodes a hypothetical protein in ''Fusobacterium peridonticum''. |
||
| Line 33: | Line 36: | ||
==References== |
==References== |
||
{{reflist}} |
|||
<references/> |
|||
[[Category:Non-coding RNA]] |
[[Category:Non-coding RNA]] |
||
Latest revision as of 21:39, 6 October 2024
| Fusobacteriales-1 | |
|---|---|
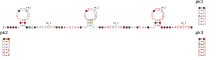 Consensus secondary structure and sequence conservation of Fusobacteriales-1 RNA | |
| Identifiers | |
| Symbol | Fusobacteriales-1 |
| Rfam | RF02920 |
| Other data | |
| RNA type | Cis-reg |
| SO | SO:0005836 |
| PDB structures | PDBe |
The Fusobacteriales-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics.[1] Fusobacteriales-1 motif RNAs are found in Fusobacteriales.
Most Fusobacteriales-1 RNAs are located upstream of genes homologous to the locus FSAG_00736, which encodes a hypothetical protein in Fusobacterium peridonticum. However, some Fusobacteriales-1 RNAs are located upstream of genes that do not appear to be homologous to FSAG_00736. Additionally 1 Fusobacteriales-1 RNAs is not found upstream of a protein-coding gene. In view of this information, it is ambiguous whether Fusobacteriales-1 RNAs function as cis-regulatory elements or whether they operate in trans.
References
[edit]- ^ Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR (October 2017). "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. doi:10.1093/nar/gkx699. PMC 5737381. PMID 28977401.
