Interferon gamma: Difference between revisions
→top: minor ce used spelt out name as per talk page |
|||
| (6 intermediate revisions by 6 users not shown) | |||
| Line 39: | Line 39: | ||
| UNII_Ref = {{fdacite|correct|FDA}} |
| UNII_Ref = {{fdacite|correct|FDA}} |
||
| UNII = 21K6M2I7AG |
| UNII = 21K6M2I7AG |
||
| KEGG = D00747 |
|||
| ATC_prefix = L03 |
| ATC_prefix = L03 |
||
| ATC_suffix = AB03 |
| ATC_suffix = AB03 |
||
| Line 50: | Line 51: | ||
| C=761 | H=1206 | N=214 | O=225 | S=6 |
| C=761 | H=1206 | N=214 | O=225 | S=6 |
||
}} |
}} |
||
'''Interferon gamma''' ('''IFNG''') is a [[protein dimer|dimer]]ized soluble [[cytokine]] that is the only member of the '''type II''' class of [[interferon]]s.<ref name="pmid6180322">{{cite journal | vauthors = Gray PW, Goeddel DV | title = Structure of the human immune interferon gene | journal = Nature | volume = 298 | issue = 5877 | pages = 859–863 | date = August 1982 | pmid = 6180322 | doi = 10.1038/298859a0 | s2cid = 4275528 | bibcode = 1982Natur.298..859G }}</ref> The existence of this interferon, which early in its history was known as immune interferon, was described by E. F. Wheelock as a product of human [[leukocyte]]s stimulated with [[phytohemagglutinin]], and by others as a product of antigen-stimulated [[lymphocytes]].<ref name="pmid17838106">{{cite journal | vauthors = Wheelock EF | title = Interferon-Like Virus-Inhibitor Induced in Human Leukocytes by Phytohemagglutinin | journal = Science | volume = 149 | issue = 3681 | pages = 310–311 | date = July 1965 | pmid = 17838106 | doi = 10.1126/science.149.3681.310 | s2cid = 1366348 | bibcode = 1965Sci...149..310W }}</ref> It was also shown to be produced in human lymphocytes.<ref>{{cite journal | vauthors = Green JA, Cooperband SR, Kibrick S | title = Immune specific induction of interferon production in cultures of human blood lymphocytes | journal = Science | volume = 164 | issue = 3886 | pages = 1415–1417 | date = June 1969 | pmid = 5783715 | doi = 10.1126/science.164.3886.1415 | s2cid = 32651832 | bibcode = 1969Sci...164.1415G }}</ref> or [[tuberculin]]-sensitized mouse [[peritoneal]] lymphocytes<ref>{{cite journal | vauthors = Milstone LM, Waksman BH | title = Release of virus inhibitor from tuberculin-sensitized peritoneal cells stimulated by antigen | journal = Journal of Immunology | volume = 105 | issue = 5 | pages = 1068–1071 | date = November 1970 | doi = 10.4049/jimmunol.105.5.1068 | pmid = 4321289 | s2cid = 29861335 | doi-access = free }}</ref> challenged with [[Mantoux test]] (PPD); the resulting [[supernatant]]s were shown to inhibit growth of [[vesicular stomatitis virus]]. Those reports also contained the basic observation underlying the now widely employed [[interferon gamma release assay]] used to test for [[tuberculosis]]. In humans, the IFNG protein is encoded by the ''IFNG'' [[gene]].<ref name="pmid6403645">{{cite journal | vauthors = Naylor SL, Sakaguchi AY, Shows TB, Law ML, Goeddel DV, Gray PW | title = Human immune interferon gene is located on chromosome 12 | journal = The Journal of Experimental Medicine | volume = 157 | issue = 3 | pages = 1020–1027 | date = March 1983 | pmid = 6403645 | pmc = 2186972 | doi = 10.1084/jem.157.3.1020 }}</ref><ref name="entrez">{{cite web | title = Entrez Gene: IFNGR2 | url = https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=3460 }}</ref> |
'''Interferon gamma''' ('''IFNG''' or IFN-γ) is a [[protein dimer|dimer]]ized soluble [[cytokine]] that is the only member of the '''type II''' class of [[interferon]]s.<ref name="pmid6180322">{{cite journal | vauthors = Gray PW, Goeddel DV | title = Structure of the human immune interferon gene | journal = Nature | volume = 298 | issue = 5877 | pages = 859–863 | date = August 1982 | pmid = 6180322 | doi = 10.1038/298859a0 | s2cid = 4275528 | bibcode = 1982Natur.298..859G }}</ref> The existence of this interferon, which early in its history was known as immune interferon, was described by E. F. Wheelock as a product of human [[leukocyte]]s stimulated with [[phytohemagglutinin]], and by others as a product of antigen-stimulated [[lymphocytes]].<ref name="pmid17838106">{{cite journal | vauthors = Wheelock EF | title = Interferon-Like Virus-Inhibitor Induced in Human Leukocytes by Phytohemagglutinin | journal = Science | volume = 149 | issue = 3681 | pages = 310–311 | date = July 1965 | pmid = 17838106 | doi = 10.1126/science.149.3681.310 | s2cid = 1366348 | bibcode = 1965Sci...149..310W }}</ref> It was also shown to be produced in human lymphocytes.<ref>{{cite journal | vauthors = Green JA, Cooperband SR, Kibrick S | title = Immune specific induction of interferon production in cultures of human blood lymphocytes | journal = Science | volume = 164 | issue = 3886 | pages = 1415–1417 | date = June 1969 | pmid = 5783715 | doi = 10.1126/science.164.3886.1415 | s2cid = 32651832 | bibcode = 1969Sci...164.1415G }}</ref> or [[tuberculin]]-sensitized mouse [[peritoneal]] lymphocytes<ref>{{cite journal | vauthors = Milstone LM, Waksman BH | title = Release of virus inhibitor from tuberculin-sensitized peritoneal cells stimulated by antigen | journal = Journal of Immunology | volume = 105 | issue = 5 | pages = 1068–1071 | date = November 1970 | doi = 10.4049/jimmunol.105.5.1068 | pmid = 4321289 | s2cid = 29861335 | doi-access = free }}</ref> challenged with [[Mantoux test]] (PPD); the resulting [[supernatant]]s were shown to inhibit growth of [[vesicular stomatitis virus]]. Those reports also contained the basic observation underlying the now widely employed [[interferon gamma release assay]] used to test for [[tuberculosis]]. In humans, the IFNG protein is encoded by the ''IFNG'' [[gene]].<ref name="pmid6403645">{{cite journal | vauthors = Naylor SL, Sakaguchi AY, Shows TB, Law ML, Goeddel DV, Gray PW | title = Human immune interferon gene is located on chromosome 12 | journal = The Journal of Experimental Medicine | volume = 157 | issue = 3 | pages = 1020–1027 | date = March 1983 | pmid = 6403645 | pmc = 2186972 | doi = 10.1084/jem.157.3.1020 }}</ref><ref name="entrez">{{cite web | title = Entrez Gene: IFNGR2 | url = https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=3460 }}</ref> |
||
Through cell signaling, interferon gamma plays a role in regulating the immune response of its target cell.<ref name=":0" /> A key signaling pathway that is activated by type II IFN is the [[JAK-STAT signaling pathway]].<ref name=":03" /> IFNG plays an important role in both [[Innate immune system|innate]] and [[Adaptive immune system|adaptive]] immunity. Type II IFN is primarily secreted by |
Through cell signaling, interferon gamma plays a role in regulating the immune response of its target cell.<ref name=":0" /> A key signaling pathway that is activated by type II IFN is the [[JAK-STAT signaling pathway]].<ref name=":03" /> IFNG plays an important role in both [[Innate immune system|innate]] and [[Adaptive immune system|adaptive]] immunity. Type II IFN is primarily secreted by CD4<sup>+</sup> [[T helper cell|T helper]] 1 (Th1) cells, [[Natural killer cell|natural killer]] (NK) cells, and CD8<sup>+</sup> [[cytotoxic T cell]]s. The expression of type II IFN is upregulated and downregulated by cytokines.<ref name=":2" /> By activating signaling pathways in cells such as [[macrophage]]s, [[B cell]]s, and [[Cytotoxic T cell|CD8<sup>+</sup> cytotoxic T cells]], it is able to promote inflammation, antiviral or antibacterial activity, and cell [[Cell proliferation|proliferation]] and [[Cellular differentiation|differentiation]].<ref name=":1" /> Type II IFN is serologically different from [[interferon type 1]], binds to different receptors, and is encoded by a separate chromosomal locus.<ref>{{cite journal | vauthors = Lee AJ, Ashkar AA | title = The Dual Nature of Type I and Type II Interferons | journal = Frontiers in Immunology | volume = 9 | pages = 2061 | date = 2018 | pmid = 30254639 | pmc = 6141705 | doi = 10.3389/fimmu.2018.02061 | doi-access = free }}</ref> Type II IFN has played a role in the development of [[cancer immunotherapy]] treatments due to its ability to prevent tumor growth.<ref name=":2" /> |
||
== Function == |
== Function == |
||
IFNG, or type II interferon, is a cytokine that is critical for [[innate immunity|innate]] and [[adaptive immunity]] against [[Viral disease|viral]], some [[Pathogenic bacteria|bacterial]] and [[protozoan infection]]s. IFNG is an important activator of [[macrophage]]s and inducer of [[major histocompatibility complex class II]] molecule expression. Aberrant IFNG expression is associated with a number of [[autoinflammatory disease|autoinflammatory]] and [[autoimmune disease]]s. The importance of IFNG in the [[immune system]] stems in part from its ability to inhibit [[viral replication]] directly, and most importantly from its [[immunostimulator]]y and [[immunomodulator]]y effects. IFNG is produced predominantly by [[natural killer cell]]s (NK) and [[natural killer T cell]]s (NKT) as part of the innate immune response, and by [[CD4]] Th1 and [[CD8]] cytotoxic T lymphocyte ([[Cytotoxic T cell|CTL]]) effector T cells once [[antigen]]-specific immunity develops<ref name="entrez2">{{cite web | title = Entrez Gene: INFG | url = https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=3458 }}</ref><ref name="pmid17981204">{{cite book | vauthors = Schoenborn JR, Wilson CB | title = Regulation of interferon-gamma during innate and adaptive immune responses | series = Advances in Immunology | volume = 96 | pages = 41–101 | year = 2007 | pmid = 17981204 | doi = 10.1016/S0065-2776(07)96002-2 | chapter = Regulation of Interferon-γ During Innate and Adaptive Immune Responses | isbn = 978-0-12-373709-0 }}</ref> as part of the adaptive immune response. IFNG is also produced by non-cytotoxic [[innate lymphoid cell]]s (ILC), a family of immune cells first discovered in the early 2010s.<ref>{{cite journal | vauthors = Artis D, Spits H | title = The biology of innate lymphoid cells | journal = Nature | volume = 517 | issue = 7534 | pages = 293–301 | date = January 2015 | pmid = 25592534 | doi = 10.1038/nature14189 | s2cid = 4386692 | bibcode = 2015Natur.517..293A }}</ref> |
|||
The primary cells that secrete type II IFN are CD4<sup>+</sup> [[T helper cell|T helper]] 1 (Th1) cells, [[Natural killer cell|natural killer]] (NK) cells, and CD8<sup>+</sup> [[cytotoxic T cell]]s. It can also be secreted by antigen presenting cells ([[Antigen-presenting cell|APCs]]) such as dendritic cells ([[Dendritic cell|DCs]]), macrophages ([[Macrophage|MΦs]]), and [[B cell]]s to a lesser degree. Type II IFN expression is upregulated by the production of [[interleukin]] cytokines, such as [[Interleukin 12|IL-12]], [[Interleukin 15|IL-15]], [[Interleukin 18|IL-18]], as well as [[Interferon type I|type I interferons]] (IFN-α and IFN-β).<ref name=":2">{{cite journal | vauthors = Castro F, Cardoso AP, Gonçalves RM, Serre K, Oliveira MJ | title = Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion | journal = Frontiers in Immunology | volume = 9 | pages = 847 | date = 2018 | pmid = 29780381 | pmc = 5945880 | doi = 10.3389/fimmu.2018.00847 | doi-access = free }}</ref> Meanwhile, [[Interleukin 4|IL-4]], [[Interleukin 10|IL-10]], transforming growth factor-beta ( |
The primary cells that secrete type II IFN are CD4<sup>+</sup> [[T helper cell|T helper]] 1 (Th1) cells, [[Natural killer cell|natural killer]] (NK) cells, and CD8<sup>+</sup> [[cytotoxic T cell]]s. It can also be secreted by antigen presenting cells ([[Antigen-presenting cell|APCs]]) such as dendritic cells ([[Dendritic cell|DCs]]), macrophages ([[Macrophage|MΦs]]), and [[B cell]]s to a lesser degree. Type II IFN expression is upregulated by the production of [[interleukin]] cytokines, such as [[Interleukin 12|IL-12]], [[Interleukin 15|IL-15]], [[Interleukin 18|IL-18]], as well as [[Interferon type I|type I interferons]] (IFN-α and IFN-β).<ref name=":2">{{cite journal | vauthors = Castro F, Cardoso AP, Gonçalves RM, Serre K, Oliveira MJ | title = Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion | journal = Frontiers in Immunology | volume = 9 | pages = 847 | date = 2018 | pmid = 29780381 | pmc = 5945880 | doi = 10.3389/fimmu.2018.00847 | doi-access = free }}</ref> Meanwhile, [[Interleukin 4|IL-4]], [[Interleukin 10|IL-10]], [[transforming growth factor-beta]] (TGF-β) and [[glucocorticoid]]s are known to downregulate type II IFN expression.<ref name=":1">{{cite journal | vauthors = Bhat MY, Solanki HS, Advani J, Khan AA, Keshava Prasad TS, Gowda H, Thiyagarajan S, Chatterjee A | title = Comprehensive network map of interferon gamma signaling | journal = Journal of Cell Communication and Signaling | volume = 12 | issue = 4 | pages = 745–751 | date = December 2018 | pmid = 30191398 | pmc = 6235777 | doi = 10.1007/s12079-018-0486-y }}</ref> |
||
Type II IFN is a cytokine, meaning it functions by signaling to other cells in the immune system and influencing their immune response. There are many immune cells type II IFN acts on. Some of its main functions are to induce [[Immunoglobulin G|IgG]] [[Immunoglobulin class switching|isotype switching]] in [[B cell]]s; upregulate [[MHC class II|major histocompatibility complex (MHC) class II]] expression on [[Antigen-presenting cell|APCs]]; induce CD8<sup>+</sup> cytotoxic T cell differentiation, activation, and proliferation; and activate [[macrophage]]s. In macrophages, type II IFN stimulates [[Interleukin 12|IL-12]] expression. IL-12 in turn promotes the secretion of |
Type II IFN is a cytokine, meaning it functions by signaling to other cells in the immune system and influencing their immune response. There are many immune cells type II IFN acts on. Some of its main functions are to induce [[Immunoglobulin G|IgG]] [[Immunoglobulin class switching|isotype switching]] in [[B cell]]s; upregulate [[MHC class II|major histocompatibility complex (MHC) class II]] expression on [[Antigen-presenting cell|APCs]]; induce CD8<sup>+</sup> cytotoxic T cell differentiation, activation, and proliferation; and activate [[macrophage]]s. In macrophages, type II IFN stimulates [[Interleukin 12|IL-12]] expression. IL-12 in turn promotes the secretion of IFNG by NK cells and Th1 cells, and it signals [[Naive T cell|naive T helper cells]] (Th0) to differentiate into Th1 cells.<ref name=":0">{{cite journal | vauthors = Tau G, Rothman P | title = Biologic functions of the IFN-gamma receptors | journal = Allergy | volume = 54 | issue = 12 | pages = 1233–1251 | date = December 1999 | pmid = 10688427 | pmc = 4154595 | doi = 10.1034/j.1398-9995.1999.00099.x }}</ref> |
||
== Structure == |
== Structure == |
||
The |
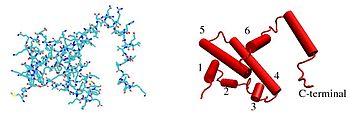
The IFNG [[monomer]] consists of a core of six α-helices and an extended unfolded sequence in the C-terminal region.<ref name="pmid1902591">{{cite journal | vauthors = Ealick SE, Cook WJ, Vijay-Kumar S, Carson M, Nagabhushan TL, Trotta PP, Bugg CE | title = Three-dimensional structure of recombinant human interferon-gamma | journal = Science | volume = 252 | issue = 5006 | pages = 698–702 | date = May 1991 | pmid = 1902591 | doi = 10.1126/science.1902591 | bibcode = 1991Sci...252..698E }}</ref><ref name="PDB_1FG9">{{PDB|1FG9}}; {{cite journal | vauthors = Thiel DJ, le Du MH, Walter RL, D'Arcy A, Chène C, Fountoulakis M, Garotta G, Winkler FK, Ealick SE | title = Observation of an unexpected third receptor molecule in the crystal structure of human interferon-gamma receptor complex | journal = Structure | volume = 8 | issue = 9 | pages = 927–936 | date = September 2000 | pmid = 10986460 | doi = 10.1016/S0969-2126(00)00184-2 | doi-access = free }}</ref> This is shown in the structural models below. The α-helices in the core of the structure are numbered 1 to 6. |
||
[[Image:IFN2.jpeg|350px|none|thumb|<span style="font-size:100%;">'''Figure 1.'''</span> Line and cartoon representation of an IFN-γ monomer.<ref name="PDB_1FG9"/>]] |
[[Image:IFN2.jpeg|350px|none|thumb|<span style="font-size:100%;">'''Figure 1.'''</span> Line and cartoon representation of an IFN-γ monomer.<ref name="PDB_1FG9"/>]] |
||
| Line 70: | Line 71: | ||
== Receptor binding == |
== Receptor binding == |
||
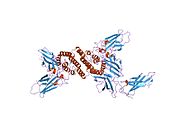
[[Image:IFN with recep.jpeg|250px|thumb|left|<span style="font-size:100%;">'''Figure 3.'''</span> IFN dimer interacting with two [[IFNGR1]] receptor molecules.<ref name="PDB_1FG9"/>]] |
[[Image:IFN with recep.jpeg|250px|thumb|left|<span style="font-size:100%;">'''Figure 3.'''</span> IFN dimer interacting with two [[IFNGR1]] receptor molecules.<ref name="PDB_1FG9"/>]] |
||
{{see also|Interferon-gamma receptor}}Cellular responses to |
{{see also|Interferon-gamma receptor}}Cellular responses to IFNG are activated through its interaction with a heterodimeric receptor consisting of [[Interferon gamma receptor 1]] (IFNGR1) and [[Interferon gamma receptor 2]] (IFNGR2). IFN-γ binding to the receptor activates the [[JAK-STAT pathway]]. Activation of the JAK-STAT pathway induces upregulation of [[interferon-stimulated gene]]s (ISGs), including MHC II.<ref>{{cite journal | vauthors = Hu X, Ivashkiv LB | title = Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases | journal = Immunity | volume = 31 | issue = 4 | pages = 539–550 | date = October 2009 | pmid = 19833085 | pmc = 2774226 | doi = 10.1016/j.immuni.2009.09.002 }}</ref> IFNG also binds to the [[glycosaminoglycan]] [[heparan sulfate]] (HS) at the cell surface. However, in contrast to many other heparan sulfate binding proteins, where binding promotes [[biological activity]], the binding of IFNG to HS inhibits its biological activity.<ref name="pmid9556569">{{cite journal | vauthors = Sadir R, Forest E, Lortat-Jacob H | title = The heparan sulfate binding sequence of interferon-gamma increased the on rate of the interferon-gamma-interferon-gamma receptor complex formation | journal = The Journal of Biological Chemistry | volume = 273 | issue = 18 | pages = 10919–10925 | date = May 1998 | pmid = 9556569 | doi = 10.1074/jbc.273.18.10919 | doi-access = free }}</ref> |
||
The structural models shown in figures 1-3 for |
The structural models shown in figures 1-3 for IFNG<ref name="PDB_1FG9"/> are all shortened at their C-termini by 17 amino acids. Full length IFNG is 143 amino acids long, the models are 126 amino acids long. Affinity for heparan sulfate resides solely within the deleted sequence of 17 amino acids.<ref name="pmid15270718">{{cite journal | vauthors = Vanhaverbeke C, Simorre JP, Sadir R, Gans P, Lortat-Jacob H | title = NMR characterization of the interaction between the C-terminal domain of interferon-gamma and heparin-derived oligosaccharides | journal = The Biochemical Journal | volume = 384 | issue = Pt 1 | pages = 93–99 | date = November 2004 | pmid = 15270718 | pmc = 1134092 | doi = 10.1042/BJ20040757 }}</ref> Within this sequence of 17 amino acids lie two clusters of basic amino acids termed D1 and D2, respectively. Heparan sulfate interacts with both of these clusters.<ref name="pmid1901275">{{cite journal | vauthors = Lortat-Jacob H, Grimaud JA | title = Interferon-gamma binds to heparan sulfate by a cluster of amino acids located in the C-terminal part of the molecule | journal = FEBS Letters | volume = 280 | issue = 1 | pages = 152–154 | date = March 1991 | pmid = 1901275 | doi = 10.1016/0014-5793(91)80225-R | s2cid = 45942972 | doi-access = free | bibcode = 1991FEBSL.280..152L }}</ref> In the absence of heparan sulfate the presence of the D1 sequence increases the rate at which IFNG-receptor complexes form.<ref name="pmid9556569"/> Interactions between the D1 cluster of amino acids and the receptor may be the first step in complex formation. By binding to D1 HS may compete with the receptor and prevent active receptor complexes from forming.{{citation needed|date=January 2023}} |
||
The biological significance of heparan sulfates interaction with |
The biological significance of heparan sulfates interaction with IFNG is unclear; however, binding of the D1 cluster to HS may protect it from [[proteolytic cleavage]].<ref name="pmid1901275"/> |
||
== Signaling == |
== Signaling == |
||
IFNG binds to the type II cell-surface receptor, also known as the IFN gamma receptor (IFNGR) which is part of the class II cytokine receptor family. The IFNGR is composed of two subunits: the [[Interferon gamma receptor 1|IFNGR1]] and [[Interferon gamma receptor 2|IFNGR2]]. IFNGR1 is associated with [[Janus kinase 1|JAK1]] and IFNGR2 is associated with [[Janus kinase 2|JAK2]]. Upon IFNG binding the receptor, IFNGR1 and IFNGR2 undergo conformational changes that result in the autophosphorylation and activation of JAK1 and JAK2. This leads to a signaling cascade and eventual transcription of target genes.<ref name=":03">{{cite journal | vauthors = Platanias LC | title = Mechanisms of type-I- and type-II-interferon-mediated signalling | journal = Nature Reviews. Immunology | volume = 5 | issue = 5 | pages = 375–386 | date = May 2005 | pmid = 15864272 | doi = 10.1038/nri1604 | s2cid = 1472195 | doi-access = free }}</ref> The expression of 236 different genes has been linked to type II IFN-mediated signaling. The proteins expressed by type II IFN-mediated signaling are primarily involved in promoting inflammatory immune responses and regulating other cell-mediated immune responses, such as [[apoptosis]], intracellular [[Immunoglobulin G|IgG]] trafficking, [[cytokine]] signaling and production, [[Haematopoiesis|hematopoiesis]], and cell [[Cell proliferation|proliferation]] and [[Cellular differentiation|differentiation]].<ref name=":1" /> |
|||
=== JAK-STAT pathway === |
=== JAK-STAT pathway === |
||
One key pathway triggered by |
One key pathway triggered by IFNG binding IFNGRs is the Janus Kinase and Signal Transducer and Activator of Transcription pathway, more commonly referred to as the [[JAK-STAT signaling pathway|JAK-STAT pathway]]. In the JAK-STAT pathway, activated JAK1 and JAK2 proteins regulate the phosphorylation of tyrosine in [[STAT1]] transcription factors. The tyrosines are phosphorylated at a very specific location, allowing activated STAT1 proteins to interact with each other come together to form STAT1-STAT1 [[Protein dimer|homodimers]]. The STAT1-STAT1 homodimers can then enter the cell nucleus. They then initiate transcription by binding to gamma interferon activation site (GAS) elements,<ref name=":03" /> which are located in the promoter region of [[Interferon-stimulated gene]]s (ISGs) that express for antiviral effector proteins, as well as positive and negative regulators of type II IFN signaling pathways.<ref>{{cite journal | vauthors = Schneider WM, Chevillotte MD, Rice CM | title = Interferon-stimulated genes: a complex web of host defenses | journal = Annual Review of Immunology | volume = 32 | issue = 1 | pages = 513–545 | date = 2014-03-21 | pmid = 24555472 | pmc = 4313732 | doi = 10.1146/annurev-immunol-032713-120231 }}</ref> |
||
[[File:Type II IFN JAK-STAT Pathway.jpg|thumb|JAK-STAT signaling pathway activated by type II IFN.]] |
[[File:Type II IFN JAK-STAT Pathway.jpg|thumb|JAK-STAT signaling pathway activated by type II IFN.]] |
||
The JAK proteins also lead to the activation of phosphatidylinositol 3-kinase ([[Phosphoinositide 3-kinase|PI3K]]). PI3K leads to the activation of protein kinase C |
The JAK proteins also lead to the activation of phosphatidylinositol 3-kinase ([[Phosphoinositide 3-kinase|PI3K]]). PI3K leads to the activation of protein kinase C delta type ([[PRKCD|PKC-δ]]) which phosphorylates the amino acid serine in STAT1 transcription factors. The phosphorylation of the serine in STAT1-STAT1 homodimers are essential for the full transcription process to occur.<ref name=":03" /> |
||
=== Other signaling pathways === |
=== Other signaling pathways === |
||
Other signaling pathways that are triggered by |
Other signaling pathways that are triggered by IFNG are the [[PI3K/AKT/mTOR pathway|mTOR signaling pathway]], the [[MAPK/ERK pathway|MAPK signaling pathway]], and the [[PI3K/AKT/mTOR pathway|PI3K/AKT signaling pathway]].<ref name=":1" /> |
||
== Biological activity == |
== Biological activity == |
||
IFNG is secreted by [[T helper cell]]s (specifically, T<sub>h</sub>1 cells), [[cytotoxic T cell]]s (T<sub>C</sub> cells), macrophages, mucosal epithelial cells and [[NK cells]]. IFNG is both an important autocrine signal for professional [[Antigen-presenting cell|APCs]] in early innate immune response, and an important paracrine signal in adaptive immune response. The expression of IFNG is induced by the cytokines IL-12, IL-15, IL-18, and type I IFN.<ref>{{cite journal | vauthors = Castro F, Cardoso AP, Gonçalves RM, Serre K, Oliveira MJ | title = Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion | journal = Frontiers in Immunology | volume = 9 | pages = 847 | date = 2018 | pmid = 29780381 | doi = 10.3389/fimmu.2018.00847 | pmc = 5945880 | doi-access = free }}</ref> IFNG is the only Type II [[interferon]] and it is [[Serology|serologically]] distinct from Type I interferons; it is acid-labile, while the type I variants are acid-stable.{{citation needed|date=January 2023}} |
|||
IFNG has antiviral, immunoregulatory, and anti-tumor properties.<ref name="pmid14525967">{{cite journal | vauthors = Schroder K, Hertzog PJ, Ravasi T, Hume DA | title = Interferon-gamma: an overview of signals, mechanisms and functions | journal = Journal of Leukocyte Biology | volume = 75 | issue = 2 | pages = 163–189 | date = February 2004 | pmid = 14525967 | doi = 10.1189/jlb.0603252 | s2cid = 15862242 | doi-access = }}</ref> It alters transcription in up to 30 genes producing a variety of physiological and cellular responses. Among the effects are: |
|||
* Promotes [[NK cell]] activity<ref>{{cite journal | vauthors = Konjević GM, Vuletić AM, Mirjačić Martinović KM, Larsen AK, Jurišić VB | title = The role of cytokines in the regulation of NK cells in the tumor environment | journal = Cytokine | volume = 117 | pages = 30–40 | date = May 2019 | pmid = 30784898 | doi = 10.1016/j.cyto.2019.02.001 | s2cid = 73482632 }}</ref> |
* Promotes [[NK cell]] activity<ref>{{cite journal | vauthors = Konjević GM, Vuletić AM, Mirjačić Martinović KM, Larsen AK, Jurišić VB | title = The role of cytokines in the regulation of NK cells in the tumor environment | journal = Cytokine | volume = 117 | pages = 30–40 | date = May 2019 | pmid = 30784898 | doi = 10.1016/j.cyto.2019.02.001 | s2cid = 73482632 }}</ref> |
||
* Increases antigen presentation and [[lysosome]] activity of [[macrophage]]s. |
* Increases antigen presentation and [[lysosome]] activity of [[macrophage]]s. |
||
| Line 100: | Line 101: | ||
* Primes alveolar [[macrophage]]s against secondary bacterial infections.<ref>{{cite journal | vauthors = Hoyer FF, Naxerova K, Schloss MJ, Hulsmans M, Nair AV, Dutta P, Calcagno DM, Herisson F, Anzai A, Sun Y, Wojtkiewicz G, Rohde D, Frodermann V, Vandoorne K, Courties G, Iwamoto Y, Garris CS, Williams DL, Breton S, Brown D, Whalen M, Libby P, Pittet MJ, King KR, Weissleder R, Swirski FK, Nahrendorf M | title = Tissue-Specific Macrophage Responses to Remote Injury Impact the Outcome of Subsequent Local Immune Challenge | journal = Immunity | volume = 51 | issue = 5 | pages = 899–914.e7 | date = November 2019 | pmid = 31732166 | pmc = 6892583 | doi = 10.1016/j.immuni.2019.10.010 }}</ref><ref>{{cite journal | vauthors = Yao Y, Jeyanathan M, Haddadi S, Barra NG, Vaseghi-Shanjani M, Damjanovic D, Lai R, Afkhami S, Chen Y, Dvorkin-Gheva A, Robbins CS, Schertzer JD, Xing Z | title = Induction of Autonomous Memory Alveolar Macrophages Requires T Cell Help and Is Critical to Trained Immunity | journal = Cell | volume = 175 | issue = 6 | pages = 1634–1650.e17 | date = November 2018 | pmid = 30433869 | doi = 10.1016/j.cell.2018.09.042 | doi-access = free }}</ref> |
* Primes alveolar [[macrophage]]s against secondary bacterial infections.<ref>{{cite journal | vauthors = Hoyer FF, Naxerova K, Schloss MJ, Hulsmans M, Nair AV, Dutta P, Calcagno DM, Herisson F, Anzai A, Sun Y, Wojtkiewicz G, Rohde D, Frodermann V, Vandoorne K, Courties G, Iwamoto Y, Garris CS, Williams DL, Breton S, Brown D, Whalen M, Libby P, Pittet MJ, King KR, Weissleder R, Swirski FK, Nahrendorf M | title = Tissue-Specific Macrophage Responses to Remote Injury Impact the Outcome of Subsequent Local Immune Challenge | journal = Immunity | volume = 51 | issue = 5 | pages = 899–914.e7 | date = November 2019 | pmid = 31732166 | pmc = 6892583 | doi = 10.1016/j.immuni.2019.10.010 }}</ref><ref>{{cite journal | vauthors = Yao Y, Jeyanathan M, Haddadi S, Barra NG, Vaseghi-Shanjani M, Damjanovic D, Lai R, Afkhami S, Chen Y, Dvorkin-Gheva A, Robbins CS, Schertzer JD, Xing Z | title = Induction of Autonomous Memory Alveolar Macrophages Requires T Cell Help and Is Critical to Trained Immunity | journal = Cell | volume = 175 | issue = 6 | pages = 1634–1650.e17 | date = November 2018 | pmid = 30433869 | doi = 10.1016/j.cell.2018.09.042 | doi-access = free }}</ref> |
||
IFNG is the primary [[cytokine]] that defines T<sub>h</sub>1 cells: T<sub>h</sub>1 cells secrete IFNG, which in turn causes more undifferentiated CD4<sup>+</sup> cells (Th0 cells) to differentiate into T<sub>h</sub>1 cells, <ref>{{cite journal | vauthors = Luckheeram RV, Zhou R, Verma AD, Xia B | title = CD4⁺T cells: differentiation and functions | journal = Clinical & Developmental Immunology | volume = 2012 | pages = 925135 | date = 2012 | pmid = 22474485 | pmc = 3312336 | doi = 10.1155/2012/925135 | doi-access = free }}</ref> representing a [[positive feedback loop]]—while suppressing T<sub>h</sub>2 cell differentiation. (Equivalent defining cytokines for other cells include [[Interleukin 4|IL-4]] for T<sub>h</sub>2 cells and [[Interleukin 17|IL-17]] for [[T helper 17 cell|Th17 cells]].) |
|||
[[NK cell]]s and [[CD8+ cytotoxic T cell]]s also produce |
[[NK cell]]s and [[CD8+ cytotoxic T cell]]s also produce IFNG. IFNG suppresses [[osteoclast]] formation by rapidly degrading the [[RANK]] adaptor protein [[TRAF6]] in the [[RANK]]-[[RANKL]] signaling pathway, which otherwise stimulates the production of [[NF-κB]].{{citation needed|date=January 2023}} |
||
===Activity in granuloma formation=== |
===Activity in granuloma formation=== |
||
A [[granuloma]] is the body's way of dealing with a substance it cannot remove or sterilize. Infectious causes of granulomas (infections are typically the most common cause of granulomas) include [[tuberculosis]], [[leprosy]], [[histoplasmosis]], [[cryptococcosis]], [[coccidioidomycosis]], [[blastomycosis]], and toxoplasmosis. Examples of non-infectious granulomatous diseases are [[sarcoidosis]], [[Crohn's disease]], [[berylliosis]], [[giant-cell arteritis]], [[granulomatosis with polyangiitis]], [[eosinophilic granulomatosis with polyangiitis]], pulmonary [[rheumatoid nodule]]s, and aspiration of food and other particulate material into the lung.<ref>{{cite journal | vauthors = Mukhopadhyay S, Farver CF, Vaszar LT, Dempsey OJ, Popper HH, Mani H, Capelozzi VL, Fukuoka J, Kerr KM, Zeren EH, Iyer VK, Tanaka T, Narde I, Nomikos A, Gumurdulu D, Arava S, Zander DS, Tazelaar HD | title = Causes of pulmonary granulomas: a retrospective study of 500 cases from seven countries | journal = Journal of Clinical Pathology | volume = 65 | issue = 1 | pages = 51–57 | date = January 2012 | pmid = 22011444 | doi = 10.1136/jclinpath-2011-200336 | s2cid = 28504428 }}</ref> The infectious pathophysiology of granulomas is discussed primarily here.{{citation needed|date=January 2023}} |
A [[granuloma]] is the body's way of dealing with a substance it cannot remove or sterilize. Infectious causes of granulomas (infections are typically the most common cause of granulomas) include [[tuberculosis]], [[leprosy]], [[histoplasmosis]], [[cryptococcosis]], [[coccidioidomycosis]], [[blastomycosis]], and toxoplasmosis. Examples of non-infectious granulomatous diseases are [[sarcoidosis]], [[Crohn's disease]], [[berylliosis]], [[giant-cell arteritis]], [[granulomatosis with polyangiitis]], [[eosinophilic granulomatosis with polyangiitis]], pulmonary [[rheumatoid nodule]]s, and aspiration of food and other particulate material into the lung.<ref>{{cite journal | vauthors = Mukhopadhyay S, Farver CF, Vaszar LT, Dempsey OJ, Popper HH, Mani H, Capelozzi VL, Fukuoka J, Kerr KM, Zeren EH, Iyer VK, Tanaka T, Narde I, Nomikos A, Gumurdulu D, Arava S, Zander DS, Tazelaar HD | title = Causes of pulmonary granulomas: a retrospective study of 500 cases from seven countries | journal = Journal of Clinical Pathology | volume = 65 | issue = 1 | pages = 51–57 | date = January 2012 | pmid = 22011444 | doi = 10.1136/jclinpath-2011-200336 | s2cid = 28504428 }}</ref> The infectious pathophysiology of granulomas is discussed primarily here.{{citation needed|date=January 2023}} |
||
The key association between |
The key association between IFNG and granulomas is that IFNG activates macrophages so that they become more powerful in killing intracellular organisms.<ref>{{cite journal | vauthors = Wu C, Xue Y, Wang P, Lin L, Liu Q, Li N, Xu J, Cao X | title = IFN-γ primes macrophage activation by increasing phosphatase and tensin homolog via downregulation of miR-3473b | journal = Journal of Immunology | volume = 193 | issue = 6 | pages = 3036–3044 | date = September 2014 | pmid = 25092892 | doi = 10.4049/jimmunol.1302379 | s2cid = 90897269 | doi-access = free }}</ref> Activation of macrophages by IFNG from T<sub>h</sub>1 helper cells in [[mycobacteria]]l infections allows the macrophages to overcome the inhibition of [[phagolysosome]] maturation caused by mycobacteria (to stay alive inside macrophages).<ref>{{cite journal | vauthors = Herbst S, Schaible UE, Schneider BE | title = Interferon gamma activated macrophages kill mycobacteria by nitric oxide induced apoptosis | journal = PLOS ONE | volume = 6 | issue = 5 | pages = e19105 | date = May 2011 | pmid = 21559306 | pmc = 3085516 | doi = 10.1371/journal.pone.0019105 | bibcode = 2011PLoSO...619105H | doi-access = free }}</ref><ref>{{cite journal | vauthors = Harris J, Master SS, De Haro SA, Delgado M, Roberts EA, Hope JC, Keane J, Deretic V | title = Th1-Th2 polarisation and autophagy in the control of intracellular mycobacteria by macrophages | journal = Veterinary Immunology and Immunopathology | volume = 128 | issue = 1–3 | pages = 37–43 | date = March 2009 | pmid = 19026454 | pmc = 2789833 | doi = 10.1016/j.vetimm.2008.10.293 }}</ref> The first steps in IFNG-induced granuloma formation are activation of T<sub>h</sub>1 helper cells by macrophages releasing [[interleukin 1|IL-1]] and [[interleukin 12|IL-12]] in the presence of intracellular pathogens, and presentation of antigens from those pathogens. Next the T<sub>h</sub>1 helper cells aggregate around the macrophages and release IFNG, which activates the macrophages. Further activation of macrophages causes a cycle of further killing of intracellular bacteria, and further presentation of antigens to T<sub>h</sub>1 helper cells with further release of IFNG. Finally, macrophages surround the T<sub>h</sub>1 helper cells and become fibroblast-like cells walling off the infection.{{citation needed|date=January 2023}} |
||
===Activity during pregnancy=== |
===Activity during pregnancy=== |
||
[[Uterine |
[[Uterine natural killer cells]] (NKs) secrete high levels of [[chemoattractant]]s, such as IFNG in mice. IFNG dilates and thins the walls of maternal spiral arteries to enhance blood flow to the [[Implantation (embryology)|implantation site]]. This remodeling aids in the development of the placenta as it invades the uterus in its quest for nutrients. IFNG knockout mice fail to initiate normal pregnancy-induced modification of [[decidual]] arteries. These models display abnormally low amounts of cells or [[necrosis]] of decidua.<ref name="pmid10899912">{{cite journal | vauthors = Ashkar AA, Di Santo JP, Croy BA | title = Interferon gamma contributes to initiation of uterine vascular modification, decidual integrity, and uterine natural killer cell maturation during normal murine pregnancy | journal = The Journal of Experimental Medicine | volume = 192 | issue = 2 | pages = 259–270 | date = July 2000 | pmid = 10899912 | pmc = 2193246 | doi = 10.1084/jem.192.2.259 }}</ref> |
||
In humans, elevated levels of IFN |
In humans, elevated levels of IFN gamma have been associated with increased risk of miscarriage. Correlation studies have observed high IFNG levels in women with a history of spontaneous miscarriage, when compared to women with no history of spontaneous miscarriage.<ref>{{cite journal | vauthors = Micallef A, Grech N, Farrugia F, Schembri-Wismayer P, Calleja-Agius J | title = The role of interferons in early pregnancy | journal = Gynecological Endocrinology | volume = 30 | issue = 1 | pages = 1–6 | date = January 2014 | pmid = 24188446 | doi = 10.3109/09513590.2012.743011 | s2cid = 207489059 }}</ref> Additionally, low-IFNG levels are associated with women who successfully carry to term. It is possible that IFNG is cytotoxic to [[trophoblast]]s, which leads to miscarriage.<ref>{{cite journal | vauthors = Berkowitz RS, Hill JA, Kurtz CB, Anderson DJ | title = Effects of products of activated leukocytes (lymphokines and monokines) on the growth of malignant trophoblast cells in vitro | journal = American Journal of Obstetrics and Gynecology | volume = 158 | issue = 1 | pages = 199–203 | date = January 1988 | pmid = 2447775 | doi = 10.1016/0002-9378(88)90810-1 }}</ref> However, causal research on the relationship between IFNG and miscarriage has not been performed due to [[Medical ethics|ethical constraints]].{{citation needed|date=January 2023}} |
||
== Production == |
== Production == |
||
Recombinant human |
Recombinant human IFNG, as an expensive biopharmaceutical, has been expressed in different expression systems including prokaryotic, protozoan, fungal (yeasts), plant, insect and mammalian cells. Human IFNG is commonly expressed in ''[[Escherichia coli]]'', marketed as ACTIMMUNE®, however, the resulting product of the prokaryotic expression system is not glycosylated with a short half-life in the bloodstream after injection; the purification process from bacterial expression system is also very costly. Other expression systems like ''[[Pichia pastoris]]'' did not show satisfactory results in terms of yields.<ref name="Razaghi A 2016">{{cite journal | vauthors = Razaghi A, Owens L, Heimann K | title = Review of the recombinant human interferon gamma as an immunotherapeutic: Impacts of production platforms and glycosylation | journal = Journal of Biotechnology | volume = 240 | pages = 48–60 | date = December 2016 | pmid = 27794496 | doi = 10.1016/j.jbiotec.2016.10.022 }}</ref><ref>{{cite journal | vauthors = Razaghi A, Tan E, Lua LH, Owens L, Karthikeyan OP, Heimann K | title = Is Pichia pastoris a realistic platform for industrial production of recombinant human interferon gamma? | journal = Biologicals | volume = 45 | pages = 52–60 | date = January 2017 | pmid = 27810255 | doi = 10.1016/j.biologicals.2016.09.015 | s2cid = 28204059 | url = https://zenodo.org/record/1312349 }}</ref> |
||
== Therapeutic use == |
== Therapeutic use == |
||
Interferon |
Interferon gamma 1b is approved by the U.S. Food and Drug Administration to treat [[chronic granulomatous disease]]<ref name="pmid1372855">{{cite journal | vauthors = Todd PA, Goa KL | title = Interferon gamma-1b. A review of its pharmacology and therapeutic potential in chronic granulomatous disease | journal = Drugs | volume = 43 | issue = 1 | pages = 111–122 | date = January 1992 | pmid = 1372855 | doi = 10.2165/00003495-199243010-00008 | s2cid = 46986837 }}</ref> (CGD) and [[osteopetrosis]].<ref name="pmid1320672">{{cite journal | vauthors = Key LL, Ries WL, Rodriguiz RM, Hatcher HC | title = Recombinant human interferon gamma therapy for osteopetrosis | journal = The Journal of Pediatrics | volume = 121 | issue = 1 | pages = 119–124 | date = July 1992 | pmid = 1320672 | doi = 10.1016/s0022-3476(05)82557-0 }}</ref> The mechanism by which IFNG benefits CGD is via enhancing the efficacy of neutrophils against catalase-positive bacteria by correcting patients' oxidative metabolism.<ref>{{cite journal | vauthors = Errante PR, Frazão JB, Condino-Neto A | title = The use of interferon-gamma therapy in chronic granulomatous disease | journal = Recent Patents on Anti-Infective Drug Discovery | volume = 3 | issue = 3 | pages = 225–230 | date = November 2008 | pmid = 18991804 | doi = 10.2174/157489108786242378 }}</ref> |
||
It was not approved to treat idiopathic pulmonary fibrosis (IPF). In 2002, the manufacturer InterMune issued a press release saying that phase III data demonstrated survival benefit in IPF and reduced mortality by 70% in patients with mild to moderate disease. The U.S. Department of Justice charged that the release contained false and misleading statements. InterMune's chief executive, Scott Harkonen, was accused of manipulating the trial data, was convicted in 2009 of wire fraud, and was sentenced to fines and community service. Harkonen appealed his conviction to the U.S. Court of Appeals for the Ninth Circuit, and lost.<ref name=Silverman_2013>{{cite journal | vauthors = Silverman E | title = Drug Marketing. The line between scientific uncertainty and promotion of snake oil | journal = BMJ | volume = 347 | pages = f5687 | date = September 2013 | pmid = 24055923 | doi = 10.1136/bmj.f5687 | s2cid = 27716008 }}</ref> Harkonen was granted a full pardon on January 20, 2021.<ref>{{cite web |title=Statement from the Press Secretary Regarding Executive Grants of Clemency |url=https://trumpwhitehouse.archives.gov/briefings-statements/statement-press-secretary-regarding-executive-grants-clemency-012021/ |via=[[NARA|National Archives]] |work=[[whitehouse.gov]] |date=January 20, 2021}}</ref> |
It was not approved to treat idiopathic pulmonary fibrosis (IPF). In 2002, the manufacturer InterMune issued a press release saying that phase III data demonstrated survival benefit in IPF and reduced mortality by 70% in patients with mild to moderate disease. The U.S. Department of Justice charged that the release contained false and misleading statements. InterMune's chief executive, Scott Harkonen, was accused of manipulating the trial data, was convicted in 2009 of wire fraud, and was sentenced to fines and community service. Harkonen appealed his conviction to the U.S. Court of Appeals for the Ninth Circuit, and lost.<ref name=Silverman_2013>{{cite journal | vauthors = Silverman E | title = Drug Marketing. The line between scientific uncertainty and promotion of snake oil | journal = BMJ | volume = 347 | pages = f5687 | date = September 2013 | pmid = 24055923 | doi = 10.1136/bmj.f5687 | s2cid = 27716008 }}</ref> Harkonen was granted a full pardon on January 20, 2021.<ref>{{cite web |title=Statement from the Press Secretary Regarding Executive Grants of Clemency |url=https://trumpwhitehouse.archives.gov/briefings-statements/statement-press-secretary-regarding-executive-grants-clemency-012021/ |via=[[NARA|National Archives]] |work=[[whitehouse.gov]] |date=January 20, 2021}}</ref> |
||
Preliminary research on the role of |
Preliminary research on the role of IFNG in treating [[Friedreich's ataxia]] (FA) conducted by [[Children's Hospital of Philadelphia]] has found no beneficial effects in short-term (< 6-months) treatment.<ref>{{cite journal | vauthors = Wells M, Seyer L, Schadt K, Lynch DR | title = IFN-γ for Friedreich ataxia: present evidence | journal = Neurodegenerative Disease Management | volume = 5 | issue = 6 | pages = 497–504 | date = December 2015 | pmid = 26634868 | doi = 10.2217/nmt.15.52 }}</ref><ref name=FA>{{cite journal | vauthors = Seyer L, Greeley N, Foerster D, Strawser C, Gelbard S, Dong Y, Schadt K, Cotticelli MG, Brocht A, Farmer J, Wilson RB, Lynch DR | title = Open-label pilot study of interferon gamma-1b in Friedreich ataxia | journal = Acta Neurologica Scandinavica | volume = 132 | issue = 1 | pages = 7–15 | date = July 2015 | pmid = 25335475 | doi = 10.1111/ane.12337 | s2cid = 207014054 | doi-access = free }}</ref><ref>{{cite journal | vauthors = Lynch DR, Hauser L, McCormick A, Wells M, Dong YN, McCormack S, Schadt K, Perlman S, Subramony SH, Mathews KD, Brocht A, Ball J, Perdok R, Grahn A, Vescio T, Sherman JW, Farmer JM | title = Randomized, double-blind, placebo-controlled study of interferon-''γ'' 1b in Friedreich Ataxia | journal = Annals of Clinical and Translational Neurology | volume = 6 | issue = 3 | pages = 546–553 | date = March 2019 | pmid = 30911578 | pmc = 6414489 | doi = 10.1002/acn3.731 }}</ref> However, researchers in Turkey have discovered significant improvements in patients' gait and stance after 6 months of treatment.<ref>{{cite journal | vauthors = Yetkİn MF, GÜltekİn M | title = Efficacy and Tolerability of Interferon Gamma in Treatment of Friedreich's Ataxia: Retrospective Study | journal = Noro Psikiyatri Arsivi | volume = 57 | issue = 4 | pages = 270–273 | date = December 2020 | pmid = 33354116 | pmc = 7735154 | doi = 10.29399/npa.25047 }}</ref> |
||
Although not officially approved, Interferon |
Although not officially approved, Interferon gamma has also been shown to be effective in treating patients with moderate to severe [[atopic dermatitis]].<ref>{{cite journal | vauthors = Akhavan A, Rudikoff D | title = Atopic dermatitis: systemic immunosuppressive therapy | journal = Seminars in Cutaneous Medicine and Surgery | volume = 27 | issue = 2 | pages = 151–155 | date = June 2008 | pmid = 18620137 | doi = 10.1016/j.sder.2008.04.004 }}</ref><ref>{{cite journal | vauthors = Schneider LC, Baz Z, Zarcone C, Zurakowski D | title = Long-term therapy with recombinant interferon-gamma (rIFN-gamma) for atopic dermatitis | journal = Annals of Allergy, Asthma & Immunology | volume = 80 | issue = 3 | pages = 263–268 | date = March 1998 | pmid = 9532976 | doi = 10.1016/S1081-1206(10)62968-7 }}</ref><ref>{{cite journal | vauthors = Hanifin JM, Schneider LC, Leung DY, Ellis CN, Jaffe HS, Izu AE, Bucalo LR, Hirabayashi SE, Tofte SJ, Cantu-Gonzales G | title = Recombinant interferon gamma therapy for atopic dermatitis | journal = Journal of the American Academy of Dermatology | volume = 28 | issue = 2 Pt 1 | pages = 189–197 | date = February 1993 | pmid = 8432915 | doi = 10.1016/0190-9622(93)70026-p }}</ref> Specifically, recombinant IFNG therapy has shown promise in patients with lowered IFNG expression, such as those with predisposition to herpes simplex virus, and pediatric patients.<ref>{{cite journal | vauthors = Brar K, Leung DY | title = Recent considerations in the use of recombinant interferon gamma for biological therapy of atopic dermatitis | journal = Expert Opinion on Biological Therapy | volume = 16 | issue = 4 | pages = 507–514 | date = 2016 | pmid = 26694988 | pmc = 4985031 | doi = 10.1517/14712598.2016.1135898 }}</ref> |
||
==Potential use in immunotherapy== |
==Potential use in immunotherapy== |
||
IFNG increases an anti-proliferative state in cancer cells, while upregulating MHC I and MHC II expression, which increases immunorecognition and removal of pathogenic cells.<ref>{{cite journal | vauthors = Kak G, Raza M, Tiwari BK | title = Interferon-gamma (IFN-γ): Exploring its implications in infectious diseases | journal = Biomolecular Concepts | volume = 9 | issue = 1 | pages = 64–79 | date = May 2018 | pmid = 29856726 | doi = 10.1515/bmc-2018-0007 | s2cid = 46922378 | doi-access = free }}</ref> IFNG also reduces metastasis in tumors by upregulating [[fibronectin]], which negatively impacts tumor architecture.<ref>{{cite journal | vauthors = Jorgovanovic D, Song M, Wang L, Zhang Y | title = Roles of IFN-γ in tumor progression and regression: a review | journal = Biomarker Research | volume = 8 | issue = 1 | pages = 49 | date = 2020-09-29 | pmid = 33005420 | pmc = 7526126 | doi = 10.1186/s40364-020-00228-x | doi-access = free }}</ref> Increased IFNG mRNA levels in tumors at diagnosis has been associated to better responses to immunotherapy.<ref>{{cite journal | vauthors = Casarrubios M, Provencio M, Nadal E, Insa A, del Rosario García-Campelo M, Lázaro-Quintela M, Dómine M, Majem M, Rodriguez-Abreu D, Martinez-Marti A, De Castro Carpeño J, Cobo M, López Vivanco G, Del Barco E, Bernabé R, Viñolas N, Barneto Aranda I, Massuti B, Sierra-Rodero B, Martinez-Toledo C, Fernández-Miranda I, Serna-Blanco R, Romero A, Calvo V, Cruz-Bermúdez A |title=Tumor microenvironment gene expression profiles associated to complete pathological response and disease progression in resectable NSCLC patients treated with neoadjuvant chemoimmunotherapy |journal=Journal for ImmunoTherapy of Cancer |date=September 2022 |volume=10 |issue=9 |pages=e005320 |doi=10.1136/jitc-2022-005320|pmid=36171009 |pmc=9528578 |hdl=2445/190198 |hdl-access=free }}</ref> |
|||
=== Cancer immunotherapy === |
=== Cancer immunotherapy === |
||
The goal of [[cancer immunotherapy]] is to trigger an immune response by the patient's immune cells to attack and kill malignant (cancer-causing) tumor cells. Type II IFN deficiency has been linked to several types of cancer, including B-cell lymphoma and lung cancer. Furthermore, it has been found that in patients receiving the drug [[durvalumab]] to treat [[Non-small-cell lung carcinoma|non-small cell lung carcinoma]] and [[transitional cell carcinoma]] had higher response rates to the drug, and the drug stunted the progression of both types of cancer for a longer duration of time. Thus, promoting the upregulation of type II IFN has been proven to be a crucial part in creating effective cancer immunotherapy treatments.<ref name=":22">{{cite journal | vauthors = Ni L, Lu J | title = Interferon gamma in cancer immunotherapy | journal = Cancer Medicine | volume = 7 | issue = 9 | pages = 4509–4516 | date = September 2018 | pmid = 30039553 | pmc = 6143921 | doi = 10.1002/cam4.1700 }}</ref> |
The goal of [[cancer immunotherapy]] is to trigger an immune response by the patient's immune cells to attack and kill malignant (cancer-causing) tumor cells. Type II IFN deficiency has been linked to several types of cancer, including B-cell lymphoma and lung cancer. Furthermore, it has been found that in patients receiving the drug [[durvalumab]] to treat [[Non-small-cell lung carcinoma|non-small cell lung carcinoma]] and [[transitional cell carcinoma]] had higher response rates to the drug, and the drug stunted the progression of both types of cancer for a longer duration of time. Thus, promoting the upregulation of type II IFN has been proven to be a crucial part in creating effective cancer immunotherapy treatments.<ref name=":22">{{cite journal | vauthors = Ni L, Lu J | title = Interferon gamma in cancer immunotherapy | journal = Cancer Medicine | volume = 7 | issue = 9 | pages = 4509–4516 | date = September 2018 | pmid = 30039553 | pmc = 6143921 | doi = 10.1002/cam4.1700 }}</ref> |
||
IFNG is not approved yet for the treatment in any [[cancer immunotherapy]]. However, improved survival was observed when IFNG was administered to patients with [[bladder carcinoma]] and [[melanoma]] cancers. The most promising result was achieved in patients with stage 2 and 3 of [[ovarian carcinoma]]. On the contrary, it was stressed: "Interferon-γ secreted by CD8-positive lymphocytes upregulates PD-L1 on ovarian cancer cells and promotes tumour growth."<ref>{{cite journal | vauthors = Abiko K, Matsumura N, Hamanishi J, Horikawa N, Murakami R, Yamaguchi K, Yoshioka Y, Baba T, Konishi I, Mandai M | title = IFN-γ from lymphocytes induces PD-L1 expression and promotes progression of ovarian cancer | journal = British Journal of Cancer | volume = 112 | issue = 9 | pages = 1501–1509 | date = April 2015 | pmid = 25867264 | pmc = 4453666 | doi = 10.1038/bjc.2015.101 }}</ref> The ''[[in vitro]]'' study of IFNG in cancer cells is more extensive and results indicate anti-proliferative activity of IFNG leading to the growth inhibition or cell death, generally induced by [[apoptosis]] but sometimes by [[autophagy]].<ref name="Razaghi A 2016" /> In addition, it has been reported that mammalian [[glycosylation]] of [[recombinant protein|recombinant]] human IFNG, expressed in [[HEK 293 cells|HEK293]], improves its therapeutic efficacy compared to the unglycosylated form that is expressed in ''[[Escherichia coli|E. coli]]''.<ref>{{cite journal | vauthors = Razaghi A, Villacrés C, Jung V, Mashkour N, Butler M, Owens L, Heimann K | title = Improved therapeutic efficacy of mammalian expressed-recombinant interferon gamma against ovarian cancer cells | journal = Experimental Cell Research | volume = 359 | issue = 1 | pages = 20–29 | date = October 2017 | pmid = 28803068 | doi = 10.1016/j.yexcr.2017.08.014 | s2cid = 12800448 }}</ref> |
|||
=== Involvement in antitumor immunity === |
=== Involvement in antitumor immunity === |
||
| Line 142: | Line 143: | ||
== Interactions == |
== Interactions == |
||
Interferon |
Interferon gamma has been shown to [[Protein-protein interaction|interact]] with [[Interferon gamma receptor 1]] and [[Interferon gamma receptor 2]].<ref name="pmid10986460">{{cite journal | vauthors = Thiel DJ, le Du MH, Walter RL, D'Arcy A, Chène C, Fountoulakis M, Garotta G, Winkler FK, Ealick SE | title = Observation of an unexpected third receptor molecule in the crystal structure of human interferon-gamma receptor complex | journal = Structure | volume = 8 | issue = 9 | pages = 927–936 | date = September 2000 | pmid = 10986460 | doi = 10.1016/S0969-2126(00)00184-2 | doi-access = free }}</ref><ref name="pmid7673114">{{cite journal | vauthors = Kotenko SV, Izotova LS, Pollack BP, Mariano TM, Donnelly RJ, Muthukumaran G, Cook JR, Garotta G, Silvennoinen O, Ihle JN | title = Interaction between the components of the interferon gamma receptor complex | journal = The Journal of Biological Chemistry | volume = 270 | issue = 36 | pages = 20915–20921 | date = September 1995 | pmid = 7673114 | doi = 10.1074/jbc.270.36.20915 | doi-access = free }}</ref> |
||
===Diseases=== |
===Diseases=== |
||
Interferon |
Interferon gamma has been shown to be a crucial player in the immune response against some intracellular pathogens, including that of [[Chagas disease]].<ref>{{cite journal | vauthors = Leon Rodriguez DA, Carmona FD, Echeverría LE, González CI, Martin J | title = IL18 Gene Variants Influence the Susceptibility to Chagas Disease | journal = PLOS Neglected Tropical Diseases | volume = 10 | issue = 3 | pages = e0004583 | date = March 2016 | pmid = 27027876 | pmc = 4814063 | doi = 10.1371/journal.pntd.0004583 | doi-access = free }}</ref> It has also been identified as having a role in seborrheic dermatitis.<ref>{{cite journal | vauthors = Trznadel-Grodzka E, Błaszkowski M, Rotsztejn H | title = Investigations of seborrheic dermatitis. Part I. The role of selected cytokines in the pathogenesis of seborrheic dermatitis | journal = Postepy Higieny I Medycyny Doswiadczalnej | volume = 66 | pages = 843–847 | date = November 2012 | pmid = 23175340 | doi = 10.5604/17322693.1019642 | doi-access = free }}</ref> |
||
IFNG has a significant anti-viral effect in [[herpes simplex virus]] I (HSV) infection. IFNG compromises the [[microtubule]]s that HSV relies upon for transport into an infected cell's nucleus, inhibiting the ability of HSV to replicate.<ref>{{cite journal | vauthors = Bigley NJ | title = Complexity of Interferon-γ Interactions with HSV-1 | journal = Frontiers in Immunology | volume = 5 | pages = 15 | date = 2014-02-06 | pmid = 24567732 | pmc = 3915238 | doi = 10.3389/fimmu.2014.00015 | doi-access = free }}</ref><ref>{{cite journal | vauthors = Sodeik B, Ebersold MW, Helenius A | title = Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus | journal = The Journal of Cell Biology | volume = 136 | issue = 5 | pages = 1007–1021 | date = March 1997 | pmid = 9060466 | pmc = 2132479 | doi = 10.1083/jcb.136.5.1007 }}</ref> Studies in mice on [[acyclovir]] resistant herpes have shown that IFNG treatment can significantly reduce herpes viral load. The mechanism by which IFNG inhibits herpes reproduction is independent of T-cells, which means that IFNG may be an effective treatment in individuals with low T-cells.<ref>{{cite journal | vauthors = Huang WY, Su YH, Yao HW, Ling P, Tung YY, Chen SH, Wang X, Chen SH | title = Beta interferon plus gamma interferon efficiently reduces acyclovir-resistant herpes simplex virus infection in mice in a T-cell-independent manner | journal = The Journal of General Virology | volume = 91 | issue = Pt 3 | pages = 591–598 | date = March 2010 | pmid = 19906941 | doi = 10.1099/vir.0.016964-0 | doi-access = free }}</ref><ref>{{cite journal | vauthors = Sainz B, Halford WP | title = Alpha/Beta interferon and gamma interferon synergize to inhibit the replication of herpes simplex virus type 1 | journal = Journal of Virology | volume = 76 | issue = 22 | pages = 11541–11550 | date = November 2002 | pmid = 12388715 | pmc = 136787 | doi = 10.1128/JVI.76.22.11541-11550.2002 }}</ref><ref>{{cite journal | vauthors = Khanna KM, Lepisto AJ, Decman V, Hendricks RL | title = Immune control of herpes simplex virus during latency | journal = Current Opinion in Immunology | volume = 16 | issue = 4 | pages = 463–469 | date = August 2004 | pmid = 15245740 | doi = 10.1016/j.coi.2004.05.003 }}</ref> |
|||
[[Chlamydia]] infection is impacted by |
[[Chlamydia]] infection is impacted by IFNG in host cells. In human epithelial cells, IFNG upregulates expression of [[indoleamine 2,3-dioxygenase]], which in turn depletes tryptophan in hosts and impedes chlamydia's reproduction.<ref>{{cite journal | vauthors = Rottenberg ME, Gigliotti-Rothfuchs A, Wigzell H | title = The role of IFN-gamma in the outcome of chlamydial infection | journal = Current Opinion in Immunology | volume = 14 | issue = 4 | pages = 444–451 | date = August 2002 | pmid = 12088678 | doi = 10.1016/s0952-7915(02)00361-8 }}</ref><ref>{{cite journal | vauthors = Taylor MW, Feng GS | title = Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism | journal = FASEB Journal | volume = 5 | issue = 11 | pages = 2516–2522 | date = August 1991 | pmid = 1907934 | doi = 10.1096/fasebj.5.11.1907934 | doi-access = free | s2cid = 25298471 }}</ref> Additionally, in rodent epithelial cells, IFNG upregulates a [[GTPase]] that inhibits chlamydial proliferation.<ref>{{cite journal | vauthors = Bernstein-Hanley I, Coers J, Balsara ZR, Taylor GA, Starnbach MN, Dietrich WF | title = The p47 GTPases Igtp and Irgb10 map to the Chlamydia trachomatis susceptibility locus Ctrq-3 and mediate cellular resistance in mice | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 103 | issue = 38 | pages = 14092–14097 | date = September 2006 | pmid = 16959883 | pmc = 1599917 | doi = 10.1073/pnas.0603338103 | bibcode = 2006PNAS..10314092B | doi-access = free }}</ref> In both the human and rodent systems, chlamydia has evolved mechanisms to circumvent the negative effects of host cell behavior.<ref>{{cite journal | vauthors = Nelson DE, Virok DP, Wood H, Roshick C, Johnson RM, Whitmire WM, Crane DD, Steele-Mortimer O, Kari L, McClarty G, Caldwell HD | title = Chlamydial IFN-gamma immune evasion is linked to host infection tropism | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 102 | issue = 30 | pages = 10658–10663 | date = July 2005 | pmid = 16020528 | pmc = 1180788 | doi = 10.1073/pnas.0504198102 | bibcode = 2005PNAS..10210658N | doi-access = free }}</ref> |
||
==Regulation== |
==Regulation== |
||
| Line 174: | Line 175: | ||
* {{PDBe-KB2|P01579|Interferon gamma}} |
* {{PDBe-KB2|P01579|Interferon gamma}} |
||
* {{MeshName|Interferon+Type+II}} |
* {{MeshName|Interferon+Type+II}} |
||
*[https://webs.iiitd.edu.in/raghava/ifnepitope2/ IFNepitope2] Prediction of IFN-gamma inducing peptides |
|||
* {{cite web |url=https://druginfo.nlm.nih.gov/drugportal/name/interferon%20type%20ii |publisher=U.S. National Library of Medicine |work=Drug Information Portal |title=Interferon type II}} |
* {{cite web |url=https://druginfo.nlm.nih.gov/drugportal/name/interferon%20type%20ii |publisher=U.S. National Library of Medicine |work=Drug Information Portal |title=Interferon type II}} |
||
Latest revision as of 07:29, 6 November 2024
| Interferon gamma | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystal structure of a biologically active single chain mutant of human interferon gamma | |||||||||
| Identifiers | |||||||||
| Symbol | IFN gamma | ||||||||
| Pfam | PF00714 | ||||||||
| Pfam clan | CL0053 | ||||||||
| InterPro | IPR002069 | ||||||||
| SCOP2 | 1rfb / SCOPe / SUPFAM | ||||||||
| |||||||||
| Clinical data | |
|---|---|
| Trade names | Actimmune |
| AHFS/Drugs.com | Monograph |
| MedlinePlus | a601152 |
| ATC code | |
| Identifiers | |
| |
| CAS Number | |
| DrugBank | |
| ChemSpider |
|
| UNII | |
| KEGG | |
| ChEMBL | |
| Chemical and physical data | |
| Formula | C761H1206N214O225S6 |
| Molar mass | 17145.65 g·mol−1 |
| | |
Interferon gamma (IFNG or IFN-γ) is a dimerized soluble cytokine that is the only member of the type II class of interferons.[5] The existence of this interferon, which early in its history was known as immune interferon, was described by E. F. Wheelock as a product of human leukocytes stimulated with phytohemagglutinin, and by others as a product of antigen-stimulated lymphocytes.[6] It was also shown to be produced in human lymphocytes.[7] or tuberculin-sensitized mouse peritoneal lymphocytes[8] challenged with Mantoux test (PPD); the resulting supernatants were shown to inhibit growth of vesicular stomatitis virus. Those reports also contained the basic observation underlying the now widely employed interferon gamma release assay used to test for tuberculosis. In humans, the IFNG protein is encoded by the IFNG gene.[9][10]
Through cell signaling, interferon gamma plays a role in regulating the immune response of its target cell.[11] A key signaling pathway that is activated by type II IFN is the JAK-STAT signaling pathway.[12] IFNG plays an important role in both innate and adaptive immunity. Type II IFN is primarily secreted by CD4+ T helper 1 (Th1) cells, natural killer (NK) cells, and CD8+ cytotoxic T cells. The expression of type II IFN is upregulated and downregulated by cytokines.[13] By activating signaling pathways in cells such as macrophages, B cells, and CD8+ cytotoxic T cells, it is able to promote inflammation, antiviral or antibacterial activity, and cell proliferation and differentiation.[14] Type II IFN is serologically different from interferon type 1, binds to different receptors, and is encoded by a separate chromosomal locus.[15] Type II IFN has played a role in the development of cancer immunotherapy treatments due to its ability to prevent tumor growth.[13]
Function
[edit]IFNG, or type II interferon, is a cytokine that is critical for innate and adaptive immunity against viral, some bacterial and protozoan infections. IFNG is an important activator of macrophages and inducer of major histocompatibility complex class II molecule expression. Aberrant IFNG expression is associated with a number of autoinflammatory and autoimmune diseases. The importance of IFNG in the immune system stems in part from its ability to inhibit viral replication directly, and most importantly from its immunostimulatory and immunomodulatory effects. IFNG is produced predominantly by natural killer cells (NK) and natural killer T cells (NKT) as part of the innate immune response, and by CD4 Th1 and CD8 cytotoxic T lymphocyte (CTL) effector T cells once antigen-specific immunity develops[16][17] as part of the adaptive immune response. IFNG is also produced by non-cytotoxic innate lymphoid cells (ILC), a family of immune cells first discovered in the early 2010s.[18]
The primary cells that secrete type II IFN are CD4+ T helper 1 (Th1) cells, natural killer (NK) cells, and CD8+ cytotoxic T cells. It can also be secreted by antigen presenting cells (APCs) such as dendritic cells (DCs), macrophages (MΦs), and B cells to a lesser degree. Type II IFN expression is upregulated by the production of interleukin cytokines, such as IL-12, IL-15, IL-18, as well as type I interferons (IFN-α and IFN-β).[13] Meanwhile, IL-4, IL-10, transforming growth factor-beta (TGF-β) and glucocorticoids are known to downregulate type II IFN expression.[14]
Type II IFN is a cytokine, meaning it functions by signaling to other cells in the immune system and influencing their immune response. There are many immune cells type II IFN acts on. Some of its main functions are to induce IgG isotype switching in B cells; upregulate major histocompatibility complex (MHC) class II expression on APCs; induce CD8+ cytotoxic T cell differentiation, activation, and proliferation; and activate macrophages. In macrophages, type II IFN stimulates IL-12 expression. IL-12 in turn promotes the secretion of IFNG by NK cells and Th1 cells, and it signals naive T helper cells (Th0) to differentiate into Th1 cells.[11]
Structure
[edit]The IFNG monomer consists of a core of six α-helices and an extended unfolded sequence in the C-terminal region.[19][20] This is shown in the structural models below. The α-helices in the core of the structure are numbered 1 to 6.
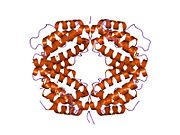
The biologically active dimer is formed by anti-parallel inter-locking of the two monomers as shown below. In the cartoon model, one monomer is shown in red, the other in blue.

Receptor binding
[edit]
Cellular responses to IFNG are activated through its interaction with a heterodimeric receptor consisting of Interferon gamma receptor 1 (IFNGR1) and Interferon gamma receptor 2 (IFNGR2). IFN-γ binding to the receptor activates the JAK-STAT pathway. Activation of the JAK-STAT pathway induces upregulation of interferon-stimulated genes (ISGs), including MHC II.[21] IFNG also binds to the glycosaminoglycan heparan sulfate (HS) at the cell surface. However, in contrast to many other heparan sulfate binding proteins, where binding promotes biological activity, the binding of IFNG to HS inhibits its biological activity.[22]
The structural models shown in figures 1-3 for IFNG[20] are all shortened at their C-termini by 17 amino acids. Full length IFNG is 143 amino acids long, the models are 126 amino acids long. Affinity for heparan sulfate resides solely within the deleted sequence of 17 amino acids.[23] Within this sequence of 17 amino acids lie two clusters of basic amino acids termed D1 and D2, respectively. Heparan sulfate interacts with both of these clusters.[24] In the absence of heparan sulfate the presence of the D1 sequence increases the rate at which IFNG-receptor complexes form.[22] Interactions between the D1 cluster of amino acids and the receptor may be the first step in complex formation. By binding to D1 HS may compete with the receptor and prevent active receptor complexes from forming.[citation needed]
The biological significance of heparan sulfates interaction with IFNG is unclear; however, binding of the D1 cluster to HS may protect it from proteolytic cleavage.[24]
Signaling
[edit]IFNG binds to the type II cell-surface receptor, also known as the IFN gamma receptor (IFNGR) which is part of the class II cytokine receptor family. The IFNGR is composed of two subunits: the IFNGR1 and IFNGR2. IFNGR1 is associated with JAK1 and IFNGR2 is associated with JAK2. Upon IFNG binding the receptor, IFNGR1 and IFNGR2 undergo conformational changes that result in the autophosphorylation and activation of JAK1 and JAK2. This leads to a signaling cascade and eventual transcription of target genes.[12] The expression of 236 different genes has been linked to type II IFN-mediated signaling. The proteins expressed by type II IFN-mediated signaling are primarily involved in promoting inflammatory immune responses and regulating other cell-mediated immune responses, such as apoptosis, intracellular IgG trafficking, cytokine signaling and production, hematopoiesis, and cell proliferation and differentiation.[14]
JAK-STAT pathway
[edit]One key pathway triggered by IFNG binding IFNGRs is the Janus Kinase and Signal Transducer and Activator of Transcription pathway, more commonly referred to as the JAK-STAT pathway. In the JAK-STAT pathway, activated JAK1 and JAK2 proteins regulate the phosphorylation of tyrosine in STAT1 transcription factors. The tyrosines are phosphorylated at a very specific location, allowing activated STAT1 proteins to interact with each other come together to form STAT1-STAT1 homodimers. The STAT1-STAT1 homodimers can then enter the cell nucleus. They then initiate transcription by binding to gamma interferon activation site (GAS) elements,[12] which are located in the promoter region of Interferon-stimulated genes (ISGs) that express for antiviral effector proteins, as well as positive and negative regulators of type II IFN signaling pathways.[25]

The JAK proteins also lead to the activation of phosphatidylinositol 3-kinase (PI3K). PI3K leads to the activation of protein kinase C delta type (PKC-δ) which phosphorylates the amino acid serine in STAT1 transcription factors. The phosphorylation of the serine in STAT1-STAT1 homodimers are essential for the full transcription process to occur.[12]
Other signaling pathways
[edit]Other signaling pathways that are triggered by IFNG are the mTOR signaling pathway, the MAPK signaling pathway, and the PI3K/AKT signaling pathway.[14]
Biological activity
[edit]IFNG is secreted by T helper cells (specifically, Th1 cells), cytotoxic T cells (TC cells), macrophages, mucosal epithelial cells and NK cells. IFNG is both an important autocrine signal for professional APCs in early innate immune response, and an important paracrine signal in adaptive immune response. The expression of IFNG is induced by the cytokines IL-12, IL-15, IL-18, and type I IFN.[26] IFNG is the only Type II interferon and it is serologically distinct from Type I interferons; it is acid-labile, while the type I variants are acid-stable.[citation needed]
IFNG has antiviral, immunoregulatory, and anti-tumor properties.[27] It alters transcription in up to 30 genes producing a variety of physiological and cellular responses. Among the effects are:
- Promotes NK cell activity[28]
- Increases antigen presentation and lysosome activity of macrophages.
- Activates inducible nitric oxide synthase (iNOS)
- Induces the production of IgG2a and IgG3 from activated plasma B cells
- Causes normal cells to increase expression of class I MHC molecules as well as class II MHC on antigen-presenting cells—to be specific, through induction of antigen processing genes, including subunits of the immunoproteasome (MECL1, LMP2, LMP7), as well as TAP and ERAAP in addition possibly to the direct upregulation of MHC heavy chains and B2-microglobulin itself
- Promotes adhesion and binding required for leukocyte migration
- Induces the expression of intrinsic defense factors—for example, with respect to retroviruses, relevant genes include TRIM5alpha, APOBEC, and Tetherin, representing directly antiviral effects
- Primes alveolar macrophages against secondary bacterial infections.[29][30]
IFNG is the primary cytokine that defines Th1 cells: Th1 cells secrete IFNG, which in turn causes more undifferentiated CD4+ cells (Th0 cells) to differentiate into Th1 cells, [31] representing a positive feedback loop—while suppressing Th2 cell differentiation. (Equivalent defining cytokines for other cells include IL-4 for Th2 cells and IL-17 for Th17 cells.)
NK cells and CD8+ cytotoxic T cells also produce IFNG. IFNG suppresses osteoclast formation by rapidly degrading the RANK adaptor protein TRAF6 in the RANK-RANKL signaling pathway, which otherwise stimulates the production of NF-κB.[citation needed]
Activity in granuloma formation
[edit]A granuloma is the body's way of dealing with a substance it cannot remove or sterilize. Infectious causes of granulomas (infections are typically the most common cause of granulomas) include tuberculosis, leprosy, histoplasmosis, cryptococcosis, coccidioidomycosis, blastomycosis, and toxoplasmosis. Examples of non-infectious granulomatous diseases are sarcoidosis, Crohn's disease, berylliosis, giant-cell arteritis, granulomatosis with polyangiitis, eosinophilic granulomatosis with polyangiitis, pulmonary rheumatoid nodules, and aspiration of food and other particulate material into the lung.[32] The infectious pathophysiology of granulomas is discussed primarily here.[citation needed]
The key association between IFNG and granulomas is that IFNG activates macrophages so that they become more powerful in killing intracellular organisms.[33] Activation of macrophages by IFNG from Th1 helper cells in mycobacterial infections allows the macrophages to overcome the inhibition of phagolysosome maturation caused by mycobacteria (to stay alive inside macrophages).[34][35] The first steps in IFNG-induced granuloma formation are activation of Th1 helper cells by macrophages releasing IL-1 and IL-12 in the presence of intracellular pathogens, and presentation of antigens from those pathogens. Next the Th1 helper cells aggregate around the macrophages and release IFNG, which activates the macrophages. Further activation of macrophages causes a cycle of further killing of intracellular bacteria, and further presentation of antigens to Th1 helper cells with further release of IFNG. Finally, macrophages surround the Th1 helper cells and become fibroblast-like cells walling off the infection.[citation needed]
Activity during pregnancy
[edit]Uterine natural killer cells (NKs) secrete high levels of chemoattractants, such as IFNG in mice. IFNG dilates and thins the walls of maternal spiral arteries to enhance blood flow to the implantation site. This remodeling aids in the development of the placenta as it invades the uterus in its quest for nutrients. IFNG knockout mice fail to initiate normal pregnancy-induced modification of decidual arteries. These models display abnormally low amounts of cells or necrosis of decidua.[36]
In humans, elevated levels of IFN gamma have been associated with increased risk of miscarriage. Correlation studies have observed high IFNG levels in women with a history of spontaneous miscarriage, when compared to women with no history of spontaneous miscarriage.[37] Additionally, low-IFNG levels are associated with women who successfully carry to term. It is possible that IFNG is cytotoxic to trophoblasts, which leads to miscarriage.[38] However, causal research on the relationship between IFNG and miscarriage has not been performed due to ethical constraints.[citation needed]
Production
[edit]Recombinant human IFNG, as an expensive biopharmaceutical, has been expressed in different expression systems including prokaryotic, protozoan, fungal (yeasts), plant, insect and mammalian cells. Human IFNG is commonly expressed in Escherichia coli, marketed as ACTIMMUNE®, however, the resulting product of the prokaryotic expression system is not glycosylated with a short half-life in the bloodstream after injection; the purification process from bacterial expression system is also very costly. Other expression systems like Pichia pastoris did not show satisfactory results in terms of yields.[39][40]
Therapeutic use
[edit]Interferon gamma 1b is approved by the U.S. Food and Drug Administration to treat chronic granulomatous disease[41] (CGD) and osteopetrosis.[42] The mechanism by which IFNG benefits CGD is via enhancing the efficacy of neutrophils against catalase-positive bacteria by correcting patients' oxidative metabolism.[43]
It was not approved to treat idiopathic pulmonary fibrosis (IPF). In 2002, the manufacturer InterMune issued a press release saying that phase III data demonstrated survival benefit in IPF and reduced mortality by 70% in patients with mild to moderate disease. The U.S. Department of Justice charged that the release contained false and misleading statements. InterMune's chief executive, Scott Harkonen, was accused of manipulating the trial data, was convicted in 2009 of wire fraud, and was sentenced to fines and community service. Harkonen appealed his conviction to the U.S. Court of Appeals for the Ninth Circuit, and lost.[44] Harkonen was granted a full pardon on January 20, 2021.[45]
Preliminary research on the role of IFNG in treating Friedreich's ataxia (FA) conducted by Children's Hospital of Philadelphia has found no beneficial effects in short-term (< 6-months) treatment.[46][47][48] However, researchers in Turkey have discovered significant improvements in patients' gait and stance after 6 months of treatment.[49]
Although not officially approved, Interferon gamma has also been shown to be effective in treating patients with moderate to severe atopic dermatitis.[50][51][52] Specifically, recombinant IFNG therapy has shown promise in patients with lowered IFNG expression, such as those with predisposition to herpes simplex virus, and pediatric patients.[53]
Potential use in immunotherapy
[edit]IFNG increases an anti-proliferative state in cancer cells, while upregulating MHC I and MHC II expression, which increases immunorecognition and removal of pathogenic cells.[54] IFNG also reduces metastasis in tumors by upregulating fibronectin, which negatively impacts tumor architecture.[55] Increased IFNG mRNA levels in tumors at diagnosis has been associated to better responses to immunotherapy.[56]
Cancer immunotherapy
[edit]The goal of cancer immunotherapy is to trigger an immune response by the patient's immune cells to attack and kill malignant (cancer-causing) tumor cells. Type II IFN deficiency has been linked to several types of cancer, including B-cell lymphoma and lung cancer. Furthermore, it has been found that in patients receiving the drug durvalumab to treat non-small cell lung carcinoma and transitional cell carcinoma had higher response rates to the drug, and the drug stunted the progression of both types of cancer for a longer duration of time. Thus, promoting the upregulation of type II IFN has been proven to be a crucial part in creating effective cancer immunotherapy treatments.[57]
IFNG is not approved yet for the treatment in any cancer immunotherapy. However, improved survival was observed when IFNG was administered to patients with bladder carcinoma and melanoma cancers. The most promising result was achieved in patients with stage 2 and 3 of ovarian carcinoma. On the contrary, it was stressed: "Interferon-γ secreted by CD8-positive lymphocytes upregulates PD-L1 on ovarian cancer cells and promotes tumour growth."[58] The in vitro study of IFNG in cancer cells is more extensive and results indicate anti-proliferative activity of IFNG leading to the growth inhibition or cell death, generally induced by apoptosis but sometimes by autophagy.[39] In addition, it has been reported that mammalian glycosylation of recombinant human IFNG, expressed in HEK293, improves its therapeutic efficacy compared to the unglycosylated form that is expressed in E. coli.[59]
Involvement in antitumor immunity
[edit]Type II IFN enhances Th1 cell, cytotoxic T cell, and APC activities, which results in an enhanced immune response against the malignant tumor cells, leading to tumor cell apoptosis and necroptosis (cell death). Furthermore, Type II IFN suppresses the activity of regulatory T cells, which are responsible for silencing immune responses against pathogens, preventing the deactivation of the immune cells involved in the killing of the tumor cells. Type II IFN prevents tumor cell division by directly acting on the tumor cells, which results in increased expression of proteins that inhibit the tumor cells from continuing through the cell cycle (i.e., cell cycle arrest). Type II IFN can also prevent tumor growth by indirectly acting on endothelial cells lining the blood vessels close to the site of the tumor, cutting off blood flow to the tumor cells and thus the supply of necessary resources for tumor cell survival and proliferation.[57]
Barriers
[edit]The importance of type II IFN in cancer immunotherapy has been acknowledged; current research is studying the effects of type II IFN on cancer, both as a solo form of treatment and as a form of treatment to be administered alongside other anticancer drugs. But type II IFN has not been approved by the Food and Drug Administration (FDA) to treat cancer, except for malignant osteoporosis. This is most likely due to the fact that while type II IFN is involved in antitumor immunity, some of its functions may enhance the progression of a cancer. When type II IFN acts on tumor cells, it may induce the expression of a transmembrane protein known as programmed death-ligand 1 (PDL1), which allows the tumor cells to evade an attack from immune cells. Type II IFN-mediated signaling may also promote angiogenesis (formation of new blood vessels to the tumor site) and tumor cell proliferation.[57]
Interactions
[edit]Interferon gamma has been shown to interact with Interferon gamma receptor 1 and Interferon gamma receptor 2.[60][61]
Diseases
[edit]Interferon gamma has been shown to be a crucial player in the immune response against some intracellular pathogens, including that of Chagas disease.[62] It has also been identified as having a role in seborrheic dermatitis.[63]
IFNG has a significant anti-viral effect in herpes simplex virus I (HSV) infection. IFNG compromises the microtubules that HSV relies upon for transport into an infected cell's nucleus, inhibiting the ability of HSV to replicate.[64][65] Studies in mice on acyclovir resistant herpes have shown that IFNG treatment can significantly reduce herpes viral load. The mechanism by which IFNG inhibits herpes reproduction is independent of T-cells, which means that IFNG may be an effective treatment in individuals with low T-cells.[66][67][68]
Chlamydia infection is impacted by IFNG in host cells. In human epithelial cells, IFNG upregulates expression of indoleamine 2,3-dioxygenase, which in turn depletes tryptophan in hosts and impedes chlamydia's reproduction.[69][70] Additionally, in rodent epithelial cells, IFNG upregulates a GTPase that inhibits chlamydial proliferation.[71] In both the human and rodent systems, chlamydia has evolved mechanisms to circumvent the negative effects of host cell behavior.[72]
Regulation
[edit]There is evidence that interferon-gamma expression is regulated by a pseudoknotted element in its 5' UTR.[73] There is also evidence that interferon-gamma is regulated either directly or indirectly by the microRNAs: miR-29.[74] Furthermore, there is evidence that interferon-gamma expression is regulated via GAPDH in T-cells. This interaction takes place in the 3'UTR, where binding of GAPDH prevents the translation of the mRNA sequence.[75]
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000111537 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000055170 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Gray PW, Goeddel DV (August 1982). "Structure of the human immune interferon gene". Nature. 298 (5877): 859–863. Bibcode:1982Natur.298..859G. doi:10.1038/298859a0. PMID 6180322. S2CID 4275528.
- ^ Wheelock EF (July 1965). "Interferon-Like Virus-Inhibitor Induced in Human Leukocytes by Phytohemagglutinin". Science. 149 (3681): 310–311. Bibcode:1965Sci...149..310W. doi:10.1126/science.149.3681.310. PMID 17838106. S2CID 1366348.
- ^ Green JA, Cooperband SR, Kibrick S (June 1969). "Immune specific induction of interferon production in cultures of human blood lymphocytes". Science. 164 (3886): 1415–1417. Bibcode:1969Sci...164.1415G. doi:10.1126/science.164.3886.1415. PMID 5783715. S2CID 32651832.
- ^ Milstone LM, Waksman BH (November 1970). "Release of virus inhibitor from tuberculin-sensitized peritoneal cells stimulated by antigen". Journal of Immunology. 105 (5): 1068–1071. doi:10.4049/jimmunol.105.5.1068. PMID 4321289. S2CID 29861335.
- ^ Naylor SL, Sakaguchi AY, Shows TB, Law ML, Goeddel DV, Gray PW (March 1983). "Human immune interferon gene is located on chromosome 12". The Journal of Experimental Medicine. 157 (3): 1020–1027. doi:10.1084/jem.157.3.1020. PMC 2186972. PMID 6403645.
- ^ "Entrez Gene: IFNGR2".
- ^ a b Tau G, Rothman P (December 1999). "Biologic functions of the IFN-gamma receptors". Allergy. 54 (12): 1233–1251. doi:10.1034/j.1398-9995.1999.00099.x. PMC 4154595. PMID 10688427.
- ^ a b c d Platanias LC (May 2005). "Mechanisms of type-I- and type-II-interferon-mediated signalling". Nature Reviews. Immunology. 5 (5): 375–386. doi:10.1038/nri1604. PMID 15864272. S2CID 1472195.
- ^ a b c Castro F, Cardoso AP, Gonçalves RM, Serre K, Oliveira MJ (2018). "Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion". Frontiers in Immunology. 9: 847. doi:10.3389/fimmu.2018.00847. PMC 5945880. PMID 29780381.
- ^ a b c d Bhat MY, Solanki HS, Advani J, Khan AA, Keshava Prasad TS, Gowda H, et al. (December 2018). "Comprehensive network map of interferon gamma signaling". Journal of Cell Communication and Signaling. 12 (4): 745–751. doi:10.1007/s12079-018-0486-y. PMC 6235777. PMID 30191398.
- ^ Lee AJ, Ashkar AA (2018). "The Dual Nature of Type I and Type II Interferons". Frontiers in Immunology. 9: 2061. doi:10.3389/fimmu.2018.02061. PMC 6141705. PMID 30254639.
- ^ "Entrez Gene: INFG".
- ^ Schoenborn JR, Wilson CB (2007). "Regulation of Interferon-γ During Innate and Adaptive Immune Responses". Regulation of interferon-gamma during innate and adaptive immune responses. Advances in Immunology. Vol. 96. pp. 41–101. doi:10.1016/S0065-2776(07)96002-2. ISBN 978-0-12-373709-0. PMID 17981204.
- ^ Artis D, Spits H (January 2015). "The biology of innate lymphoid cells". Nature. 517 (7534): 293–301. Bibcode:2015Natur.517..293A. doi:10.1038/nature14189. PMID 25592534. S2CID 4386692.
- ^ Ealick SE, Cook WJ, Vijay-Kumar S, Carson M, Nagabhushan TL, Trotta PP, et al. (May 1991). "Three-dimensional structure of recombinant human interferon-gamma". Science. 252 (5006): 698–702. Bibcode:1991Sci...252..698E. doi:10.1126/science.1902591. PMID 1902591.
- ^ a b c d e PDB: 1FG9; Thiel DJ, le Du MH, Walter RL, D'Arcy A, Chène C, Fountoulakis M, et al. (September 2000). "Observation of an unexpected third receptor molecule in the crystal structure of human interferon-gamma receptor complex". Structure. 8 (9): 927–936. doi:10.1016/S0969-2126(00)00184-2. PMID 10986460.
- ^ Hu X, Ivashkiv LB (October 2009). "Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases". Immunity. 31 (4): 539–550. doi:10.1016/j.immuni.2009.09.002. PMC 2774226. PMID 19833085.
- ^ a b Sadir R, Forest E, Lortat-Jacob H (May 1998). "The heparan sulfate binding sequence of interferon-gamma increased the on rate of the interferon-gamma-interferon-gamma receptor complex formation". The Journal of Biological Chemistry. 273 (18): 10919–10925. doi:10.1074/jbc.273.18.10919. PMID 9556569.
- ^ Vanhaverbeke C, Simorre JP, Sadir R, Gans P, Lortat-Jacob H (November 2004). "NMR characterization of the interaction between the C-terminal domain of interferon-gamma and heparin-derived oligosaccharides". The Biochemical Journal. 384 (Pt 1): 93–99. doi:10.1042/BJ20040757. PMC 1134092. PMID 15270718.
- ^ a b Lortat-Jacob H, Grimaud JA (March 1991). "Interferon-gamma binds to heparan sulfate by a cluster of amino acids located in the C-terminal part of the molecule". FEBS Letters. 280 (1): 152–154. Bibcode:1991FEBSL.280..152L. doi:10.1016/0014-5793(91)80225-R. PMID 1901275. S2CID 45942972.
- ^ Schneider WM, Chevillotte MD, Rice CM (2014-03-21). "Interferon-stimulated genes: a complex web of host defenses". Annual Review of Immunology. 32 (1): 513–545. doi:10.1146/annurev-immunol-032713-120231. PMC 4313732. PMID 24555472.
- ^ Castro F, Cardoso AP, Gonçalves RM, Serre K, Oliveira MJ (2018). "Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion". Frontiers in Immunology. 9: 847. doi:10.3389/fimmu.2018.00847. PMC 5945880. PMID 29780381.
- ^ Schroder K, Hertzog PJ, Ravasi T, Hume DA (February 2004). "Interferon-gamma: an overview of signals, mechanisms and functions". Journal of Leukocyte Biology. 75 (2): 163–189. doi:10.1189/jlb.0603252. PMID 14525967. S2CID 15862242.
- ^ Konjević GM, Vuletić AM, Mirjačić Martinović KM, Larsen AK, Jurišić VB (May 2019). "The role of cytokines in the regulation of NK cells in the tumor environment". Cytokine. 117: 30–40. doi:10.1016/j.cyto.2019.02.001. PMID 30784898. S2CID 73482632.
- ^ Hoyer FF, Naxerova K, Schloss MJ, Hulsmans M, Nair AV, Dutta P, et al. (November 2019). "Tissue-Specific Macrophage Responses to Remote Injury Impact the Outcome of Subsequent Local Immune Challenge". Immunity. 51 (5): 899–914.e7. doi:10.1016/j.immuni.2019.10.010. PMC 6892583. PMID 31732166.
- ^ Yao Y, Jeyanathan M, Haddadi S, Barra NG, Vaseghi-Shanjani M, Damjanovic D, et al. (November 2018). "Induction of Autonomous Memory Alveolar Macrophages Requires T Cell Help and Is Critical to Trained Immunity". Cell. 175 (6): 1634–1650.e17. doi:10.1016/j.cell.2018.09.042. PMID 30433869.
- ^ Luckheeram RV, Zhou R, Verma AD, Xia B (2012). "CD4⁺T cells: differentiation and functions". Clinical & Developmental Immunology. 2012: 925135. doi:10.1155/2012/925135. PMC 3312336. PMID 22474485.
- ^ Mukhopadhyay S, Farver CF, Vaszar LT, Dempsey OJ, Popper HH, Mani H, et al. (January 2012). "Causes of pulmonary granulomas: a retrospective study of 500 cases from seven countries". Journal of Clinical Pathology. 65 (1): 51–57. doi:10.1136/jclinpath-2011-200336. PMID 22011444. S2CID 28504428.
- ^ Wu C, Xue Y, Wang P, Lin L, Liu Q, Li N, et al. (September 2014). "IFN-γ primes macrophage activation by increasing phosphatase and tensin homolog via downregulation of miR-3473b". Journal of Immunology. 193 (6): 3036–3044. doi:10.4049/jimmunol.1302379. PMID 25092892. S2CID 90897269.
- ^ Herbst S, Schaible UE, Schneider BE (May 2011). "Interferon gamma activated macrophages kill mycobacteria by nitric oxide induced apoptosis". PLOS ONE. 6 (5): e19105. Bibcode:2011PLoSO...619105H. doi:10.1371/journal.pone.0019105. PMC 3085516. PMID 21559306.
- ^ Harris J, Master SS, De Haro SA, Delgado M, Roberts EA, Hope JC, et al. (March 2009). "Th1-Th2 polarisation and autophagy in the control of intracellular mycobacteria by macrophages". Veterinary Immunology and Immunopathology. 128 (1–3): 37–43. doi:10.1016/j.vetimm.2008.10.293. PMC 2789833. PMID 19026454.
- ^ Ashkar AA, Di Santo JP, Croy BA (July 2000). "Interferon gamma contributes to initiation of uterine vascular modification, decidual integrity, and uterine natural killer cell maturation during normal murine pregnancy". The Journal of Experimental Medicine. 192 (2): 259–270. doi:10.1084/jem.192.2.259. PMC 2193246. PMID 10899912.
- ^ Micallef A, Grech N, Farrugia F, Schembri-Wismayer P, Calleja-Agius J (January 2014). "The role of interferons in early pregnancy". Gynecological Endocrinology. 30 (1): 1–6. doi:10.3109/09513590.2012.743011. PMID 24188446. S2CID 207489059.
- ^ Berkowitz RS, Hill JA, Kurtz CB, Anderson DJ (January 1988). "Effects of products of activated leukocytes (lymphokines and monokines) on the growth of malignant trophoblast cells in vitro". American Journal of Obstetrics and Gynecology. 158 (1): 199–203. doi:10.1016/0002-9378(88)90810-1. PMID 2447775.
- ^ a b Razaghi A, Owens L, Heimann K (December 2016). "Review of the recombinant human interferon gamma as an immunotherapeutic: Impacts of production platforms and glycosylation". Journal of Biotechnology. 240: 48–60. doi:10.1016/j.jbiotec.2016.10.022. PMID 27794496.
- ^ Razaghi A, Tan E, Lua LH, Owens L, Karthikeyan OP, Heimann K (January 2017). "Is Pichia pastoris a realistic platform for industrial production of recombinant human interferon gamma?". Biologicals. 45: 52–60. doi:10.1016/j.biologicals.2016.09.015. PMID 27810255. S2CID 28204059.
- ^ Todd PA, Goa KL (January 1992). "Interferon gamma-1b. A review of its pharmacology and therapeutic potential in chronic granulomatous disease". Drugs. 43 (1): 111–122. doi:10.2165/00003495-199243010-00008. PMID 1372855. S2CID 46986837.
- ^ Key LL, Ries WL, Rodriguiz RM, Hatcher HC (July 1992). "Recombinant human interferon gamma therapy for osteopetrosis". The Journal of Pediatrics. 121 (1): 119–124. doi:10.1016/s0022-3476(05)82557-0. PMID 1320672.
- ^ Errante PR, Frazão JB, Condino-Neto A (November 2008). "The use of interferon-gamma therapy in chronic granulomatous disease". Recent Patents on Anti-Infective Drug Discovery. 3 (3): 225–230. doi:10.2174/157489108786242378. PMID 18991804.
- ^ Silverman E (September 2013). "Drug Marketing. The line between scientific uncertainty and promotion of snake oil". BMJ. 347: f5687. doi:10.1136/bmj.f5687. PMID 24055923. S2CID 27716008.
- ^ "Statement from the Press Secretary Regarding Executive Grants of Clemency". whitehouse.gov. January 20, 2021 – via National Archives.
- ^ Wells M, Seyer L, Schadt K, Lynch DR (December 2015). "IFN-γ for Friedreich ataxia: present evidence". Neurodegenerative Disease Management. 5 (6): 497–504. doi:10.2217/nmt.15.52. PMID 26634868.
- ^ Seyer L, Greeley N, Foerster D, Strawser C, Gelbard S, Dong Y, et al. (July 2015). "Open-label pilot study of interferon gamma-1b in Friedreich ataxia". Acta Neurologica Scandinavica. 132 (1): 7–15. doi:10.1111/ane.12337. PMID 25335475. S2CID 207014054.
- ^ Lynch DR, Hauser L, McCormick A, Wells M, Dong YN, McCormack S, et al. (March 2019). "Randomized, double-blind, placebo-controlled study of interferon-γ 1b in Friedreich Ataxia". Annals of Clinical and Translational Neurology. 6 (3): 546–553. doi:10.1002/acn3.731. PMC 6414489. PMID 30911578.
- ^ Yetkİn MF, GÜltekİn M (December 2020). "Efficacy and Tolerability of Interferon Gamma in Treatment of Friedreich's Ataxia: Retrospective Study". Noro Psikiyatri Arsivi. 57 (4): 270–273. doi:10.29399/npa.25047. PMC 7735154. PMID 33354116.
- ^ Akhavan A, Rudikoff D (June 2008). "Atopic dermatitis: systemic immunosuppressive therapy". Seminars in Cutaneous Medicine and Surgery. 27 (2): 151–155. doi:10.1016/j.sder.2008.04.004. PMID 18620137.
- ^ Schneider LC, Baz Z, Zarcone C, Zurakowski D (March 1998). "Long-term therapy with recombinant interferon-gamma (rIFN-gamma) for atopic dermatitis". Annals of Allergy, Asthma & Immunology. 80 (3): 263–268. doi:10.1016/S1081-1206(10)62968-7. PMID 9532976.
- ^ Hanifin JM, Schneider LC, Leung DY, Ellis CN, Jaffe HS, Izu AE, et al. (February 1993). "Recombinant interferon gamma therapy for atopic dermatitis". Journal of the American Academy of Dermatology. 28 (2 Pt 1): 189–197. doi:10.1016/0190-9622(93)70026-p. PMID 8432915.
- ^ Brar K, Leung DY (2016). "Recent considerations in the use of recombinant interferon gamma for biological therapy of atopic dermatitis". Expert Opinion on Biological Therapy. 16 (4): 507–514. doi:10.1517/14712598.2016.1135898. PMC 4985031. PMID 26694988.
- ^ Kak G, Raza M, Tiwari BK (May 2018). "Interferon-gamma (IFN-γ): Exploring its implications in infectious diseases". Biomolecular Concepts. 9 (1): 64–79. doi:10.1515/bmc-2018-0007. PMID 29856726. S2CID 46922378.
- ^ Jorgovanovic D, Song M, Wang L, Zhang Y (2020-09-29). "Roles of IFN-γ in tumor progression and regression: a review". Biomarker Research. 8 (1): 49. doi:10.1186/s40364-020-00228-x. PMC 7526126. PMID 33005420.
- ^ Casarrubios M, Provencio M, Nadal E, Insa A, del Rosario García-Campelo M, Lázaro-Quintela M, et al. (September 2022). "Tumor microenvironment gene expression profiles associated to complete pathological response and disease progression in resectable NSCLC patients treated with neoadjuvant chemoimmunotherapy". Journal for ImmunoTherapy of Cancer. 10 (9): e005320. doi:10.1136/jitc-2022-005320. hdl:2445/190198. PMC 9528578. PMID 36171009.
- ^ a b c Ni L, Lu J (September 2018). "Interferon gamma in cancer immunotherapy". Cancer Medicine. 7 (9): 4509–4516. doi:10.1002/cam4.1700. PMC 6143921. PMID 30039553.
- ^ Abiko K, Matsumura N, Hamanishi J, Horikawa N, Murakami R, Yamaguchi K, et al. (April 2015). "IFN-γ from lymphocytes induces PD-L1 expression and promotes progression of ovarian cancer". British Journal of Cancer. 112 (9): 1501–1509. doi:10.1038/bjc.2015.101. PMC 4453666. PMID 25867264.
- ^ Razaghi A, Villacrés C, Jung V, Mashkour N, Butler M, Owens L, et al. (October 2017). "Improved therapeutic efficacy of mammalian expressed-recombinant interferon gamma against ovarian cancer cells". Experimental Cell Research. 359 (1): 20–29. doi:10.1016/j.yexcr.2017.08.014. PMID 28803068. S2CID 12800448.
- ^ Thiel DJ, le Du MH, Walter RL, D'Arcy A, Chène C, Fountoulakis M, et al. (September 2000). "Observation of an unexpected third receptor molecule in the crystal structure of human interferon-gamma receptor complex". Structure. 8 (9): 927–936. doi:10.1016/S0969-2126(00)00184-2. PMID 10986460.
- ^ Kotenko SV, Izotova LS, Pollack BP, Mariano TM, Donnelly RJ, Muthukumaran G, et al. (September 1995). "Interaction between the components of the interferon gamma receptor complex". The Journal of Biological Chemistry. 270 (36): 20915–20921. doi:10.1074/jbc.270.36.20915. PMID 7673114.
- ^ Leon Rodriguez DA, Carmona FD, Echeverría LE, González CI, Martin J (March 2016). "IL18 Gene Variants Influence the Susceptibility to Chagas Disease". PLOS Neglected Tropical Diseases. 10 (3): e0004583. doi:10.1371/journal.pntd.0004583. PMC 4814063. PMID 27027876.
- ^ Trznadel-Grodzka E, Błaszkowski M, Rotsztejn H (November 2012). "Investigations of seborrheic dermatitis. Part I. The role of selected cytokines in the pathogenesis of seborrheic dermatitis". Postepy Higieny I Medycyny Doswiadczalnej. 66: 843–847. doi:10.5604/17322693.1019642. PMID 23175340.
- ^ Bigley NJ (2014-02-06). "Complexity of Interferon-γ Interactions with HSV-1". Frontiers in Immunology. 5: 15. doi:10.3389/fimmu.2014.00015. PMC 3915238. PMID 24567732.
- ^ Sodeik B, Ebersold MW, Helenius A (March 1997). "Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus". The Journal of Cell Biology. 136 (5): 1007–1021. doi:10.1083/jcb.136.5.1007. PMC 2132479. PMID 9060466.
- ^ Huang WY, Su YH, Yao HW, Ling P, Tung YY, Chen SH, et al. (March 2010). "Beta interferon plus gamma interferon efficiently reduces acyclovir-resistant herpes simplex virus infection in mice in a T-cell-independent manner". The Journal of General Virology. 91 (Pt 3): 591–598. doi:10.1099/vir.0.016964-0. PMID 19906941.
- ^ Sainz B, Halford WP (November 2002). "Alpha/Beta interferon and gamma interferon synergize to inhibit the replication of herpes simplex virus type 1". Journal of Virology. 76 (22): 11541–11550. doi:10.1128/JVI.76.22.11541-11550.2002. PMC 136787. PMID 12388715.
- ^ Khanna KM, Lepisto AJ, Decman V, Hendricks RL (August 2004). "Immune control of herpes simplex virus during latency". Current Opinion in Immunology. 16 (4): 463–469. doi:10.1016/j.coi.2004.05.003. PMID 15245740.
- ^ Rottenberg ME, Gigliotti-Rothfuchs A, Wigzell H (August 2002). "The role of IFN-gamma in the outcome of chlamydial infection". Current Opinion in Immunology. 14 (4): 444–451. doi:10.1016/s0952-7915(02)00361-8. PMID 12088678.
- ^ Taylor MW, Feng GS (August 1991). "Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism". FASEB Journal. 5 (11): 2516–2522. doi:10.1096/fasebj.5.11.1907934. PMID 1907934. S2CID 25298471.
- ^ Bernstein-Hanley I, Coers J, Balsara ZR, Taylor GA, Starnbach MN, Dietrich WF (September 2006). "The p47 GTPases Igtp and Irgb10 map to the Chlamydia trachomatis susceptibility locus Ctrq-3 and mediate cellular resistance in mice". Proceedings of the National Academy of Sciences of the United States of America. 103 (38): 14092–14097. Bibcode:2006PNAS..10314092B. doi:10.1073/pnas.0603338103. PMC 1599917. PMID 16959883.
- ^ Nelson DE, Virok DP, Wood H, Roshick C, Johnson RM, Whitmire WM, et al. (July 2005). "Chlamydial IFN-gamma immune evasion is linked to host infection tropism". Proceedings of the National Academy of Sciences of the United States of America. 102 (30): 10658–10663. Bibcode:2005PNAS..10210658N. doi:10.1073/pnas.0504198102. PMC 1180788. PMID 16020528.
- ^ Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R (January 2002). "Human interferon-gamma mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR". Cell. 108 (2): 221–232. doi:10.1016/S0092-8674(02)00616-5. PMID 11832212. S2CID 14722737.
- ^ Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB (April 2008). "MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components". Molecular Immunology. 45 (7): 1995–2006. doi:10.1016/j.molimm.2007.10.035. PMC 2678893. PMID 18061676.
- ^ Chang CH, Curtis JD, Maggi LB, Faubert B, Villarino AV, O'Sullivan D, et al. (June 2013). "Posttranscriptional control of T cell effector function by aerobic glycolysis". Cell. 153 (6): 1239–1251. doi:10.1016/j.cell.2013.05.016. PMC 3804311. PMID 23746840.
Further reading
[edit]- Hall SK (1997). A commotion in the blood: life, death, and the immune system. New York: Henry Holt. ISBN 978-0-8050-5841-3.
- Ikeda H, Old LJ, Schreiber RD (April 2002). "The roles of IFN gamma in protection against tumor development and cancer immunoediting". Cytokine & Growth Factor Reviews. 13 (2): 95–109. doi:10.1016/S1359-6101(01)00038-7. PMID 11900986.
- Chesler DA, Reiss CS (December 2002). "The role of IFN-gamma in immune responses to viral infections of the central nervous system". Cytokine & Growth Factor Reviews. 13 (6): 441–454. doi:10.1016/S1359-6101(02)00044-8. PMID 12401479.
- Dessein A, Kouriba B, Eboumbou C, Dessein H, Argiro L, Marquet S, et al. (October 2004). "Interleukin-13 in the skin and interferon-gamma in the liver are key players in immune protection in human schistosomiasis". Immunological Reviews. 201: 180–190. doi:10.1111/j.0105-2896.2004.00195.x. PMID 15361241. S2CID 25378236.
- Joseph AM, Kumar M, Mitra D (January 2005). "Nef: "necessary and enforcing factor" in HIV infection". Current HIV Research. 3 (1): 87–94. doi:10.2174/1570162052773013. PMID 15638726.
- Copeland KF (December 2005). "Modulation of HIV-1 transcription by cytokines and chemokines". Mini Reviews in Medicinal Chemistry. 5 (12): 1093–1101. doi:10.2174/138955705774933383. PMID 16375755.
- Chiba H, Kojima T, Osanai M, Sawada N (January 2006). "The significance of interferon-gamma-triggered internalization of tight-junction proteins in inflammatory bowel disease". Science's STKE. 2006 (316): pe1. doi:10.1126/stke.3162006pe1. PMID 16391178. S2CID 85320208.
- Tellides G, Pober JS (March 2007). "Interferon-gamma axis in graft arteriosclerosis". Circulation Research. 100 (5): 622–632. CiteSeerX 10.1.1.495.2743. doi:10.1161/01.RES.0000258861.72279.29. PMID 17363708. S2CID 254247.
External links
[edit]- Overview of all the structural information available in the PDB for UniProt: P01579 (Interferon gamma) at the PDBe-KB.
- Interferon+Type+II at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- IFNepitope2 Prediction of IFN-gamma inducing peptides
- "Interferon type II". Drug Information Portal. U.S. National Library of Medicine.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.