Antimicrobial resistance: Difference between revisions
→Development of new drugs: fix name (see GARDP's own websitee and many other sources) |
|||
| Line 1: | Line 1: | ||
{{Short description|Resistance of microbes to drugs directed against them}} |
|||
'''Antibiotic resistance''' is a type of [[drug resistance]] where a [[microorganism]] is able to survive exposure to an [[antibiotic]]. [[Gene]]s can be [[Horizontal gene transfer|transferred between bacteria in a horizontal]] fashion by [[bacterial conjugation|conjugation]], [[Transduction (genetics)|transduction]], or [[transformation (genetics)|transformation]]. Thus a gene for antibiotic resistance which had evolved via [[natural selection]] may be shared. Evolutionary stress such as exposure to antibiotics then selects for the antibiotic resistant trait. Many antibiotic resistance genes reside on [[plasmids]], facilitating their transfer. If a bacterium carries several resistance genes, it is called multiresistant or, informally, a '''superbug''' or '''super bacteria'''. |
|||
{{cs1 config|mode=cs1|name-list-style=vanc|display-authors=6}} |
|||
{{Use dmy dates|date=January 2020}} |
|||
[[File:Antibiotic sensitivity and resistance.jpg|thumb|upright=1.4|[[Disk diffusion test|Antibiotic resistance tests]]: Bacteria are streaked on dishes with white disks, each impregnated with a different antibiotic. Clear rings, such as those on the left, show that bacteria have not grown—indicating that these bacteria are not resistant. The bacteria on the right are fully resistant to three of seven and partially resistant to two of seven antibiotics tested.<ref>[http://www.microbelibrary.org/component/resource/laboratory-test/3189-kirby-bauer-disk-diffusion-susceptibility-test-protocol Kirby-Bauer Disk Diffusion Susceptibility Test Protocol] {{webarchive|url=https://web.archive.org/web/20110626190940/http://www.microbelibrary.org/component/resource/laboratory-test/3189-kirby-bauer-disk-diffusion-susceptibility-test-protocol |date=26 June 2011 }}, Jan Hudzicki, ASM</ref>|alt=Two petri dishes with antibiotic resistance tests]] |
|||
'''Antimicrobial resistance''' ('''AMR''' or '''AR''') occurs when [[microbe]]s evolve mechanisms that protect them from [[antimicrobials]], which are drugs used to treat [[Infection|infections]].<ref name="CDC About Antimicrobial Resistance">{{cite web|url=https://www.cdc.gov/antimicrobial-resistance/about/|title=About Antimicrobial Resistance|website=US [[Centers for Disease Control and Prevention]]|date=22 April 2024|access-date=11 October 2024}}</ref> This resistance affects all classes of microbes, including [[bacteria]] ([[antibiotic]] resistance), [[viruses]] ([[antiviral]] resistance), [[protozoa]] ([[antiprotozoal]] resistance), and [[fungi]] ([[antifungal]] resistance). Together, these adaptations fall under the AMR umbrella, posing significant challenges to healthcare worldwide.<ref name="WHO2014">{{cite web|title=Antimicrobial resistance Fact sheet N°194|url=https://www.who.int/mediacentre/factsheets/fs194/en/|website=who.int|access-date=7 March 2015|date=April 2014|archive-url=https://web.archive.org/web/20150310081111/http://www.who.int/mediacentre/factsheets/fs194/en/|archive-date=10 March 2015|url-status=live}}</ref> Misuse and improper management of antimicrobials are primary drivers of this resistance, though it can also occur naturally through genetic mutations and the spread of resistant genes.<ref name="Tanwar_2014">{{cite journal | vauthors = Tanwar J, Das S, Fatima Z, Hameed S | title = Multidrug resistance: an emerging crisis | journal = Interdisciplinary Perspectives on Infectious Diseases | volume = 2014 | pages = 541340 | date = 2014 | pmid = 25140175 | pmc = 4124702 | doi = 10.1155/2014/541340 | doi-access = free }}</ref> |
|||
The primary cause of antibiotic resistance is antibiotic use both within medicine and veterinary medicine. The greater the duration of exposure the greater the risk of the development of resistance irrespective of the severity of the need for antibiotics. |
|||
Microbes resistant to multiple drugs are termed '''multidrug-resistant''' (MDR) and are sometimes called '''superbugs'''.<ref name="Magiorakos">{{cite journal | vauthors = Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL | title = Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance | journal = Clinical Microbiology and Infection | volume = 18 | issue = 3 | pages = 268–281 | date = March 2012 | pmid = 21793988 | doi = 10.1111/j.1469-0691.2011.03570.x | doi-access = free }}</ref> Antibiotic resistance, a significant AMR subset, enables [[bacteria]] to survive [[antibiotic]] treatment, complicating infection management and treatment options.<ref name="WHO2014" /> Resistance arises through spontaneous mutation, [[horizontal gene transfer]], and increased selective pressure from antibiotic overuse, both in medicine and agriculture, which accelerates resistance development.<ref name=":3">{{cite journal | vauthors = Dabour R, Meirson T, Samson AO | title = Global antibiotic resistance is mostly periodic | journal = Journal of Global Antimicrobial Resistance | volume = 7 | pages = 132–134 | date = December 2016 | pmid = 27788414 | doi = 10.1016/j.jgar.2016.09.003 }}</ref> |
|||
==Causes== |
|||
{{See also|Antibiotic misuse}} |
|||
The widespread use of antibiotics both inside and outside of medicine is playing a significant role in the emergence of resistant bacteria.<ref>{{cite journal |author=Goossens H, Ferech M, Vander Stichele R, Elseviers M |title=Outpatient antibiotic use in Europe and association with resistance: a cross-national database study |journal=Lancet |volume=365 |issue=9459 |pages=579–87 |year=2005 |pmid=15708101 |doi=10.1016/S0140-6736(05)17907-0 |url=}}</ref> Antibiotics are often used in rearing animals for food and this use among others leads to the creation of resistant strains of bacteria. In some countries antibiotics are sold over the counter without a prescription which also leads to the creation of resistant strains. In supposedly well-regulated human medicine the major problem of the emergence of resistant bacteria is due to misuse and overuse of antibiotics by doctors as well as patients.<ref>{{cite web |url=http://www.who.int/mediacentre/factsheets/fs268/en/index.html |title=Use of antimicrobials outside human medicine and resultant antimicrobial resistance in humans |author=WHO |authorlink=World Health Organisation |publisher=World Health Organization |month=January |year=2002 }}</ref> Other practices contributing towards resistance include the adugmkghdition of antibiotics to the feed of livestock.<ref>{{Cite journal | doi = 10.1126/science.295.5552.27a | journal = Science | date = 4 January 2002 | volume = 295 | issue = 5552 | pages = 27–28 | author = Dan Ferber | pmid = 11778017 | title = Livestock Feed Ban Preserves Drugs' Power }}</ref><ref name="mathew"/> Household use of antibacterials in soaps and other products, although not clearly contributing to resistance, is also discouraged (as not being effective at infection control).<ref>[http://www.cdc.gov/getsmart/antibiotic-use/anitbiotic-resistance-faqs.html#j "Are antibacterial-containing products (soaps, household cleaners, etc.) better for preventing the spread of infection? Does their use add to the problem of resistance?"], ''Antibiotic Resistance Questions & Answers'', Centers for Disease Control and Prevention, Atlanta, Georgia, USA, accessed November 17, 2009</ref> Also unsound practices in the pharmaceutical manufacturing industry can contribute towards the likelihood of creating antibiotic resistant strains.<ref>{{Cite journal | last1 = Larsson | first1 = DG. | last2 = Fick | first2 = J. | title = Transparency throughout the production chain -- a way to reduce pollution from the manufacturing of pharmaceuticals? | journal = Regul Toxicol Pharmacol | month = Jan | year = 2009 | doi = 10.1016/j.yrtph.2009.01.008 | pmid = 19545507 | volume = 53 | pages = 161 | issue = 3 }}</ref> |
|||
The burden of AMR is immense, with nearly 5 million annual deaths associated with resistant infections.<ref name=WHO10October2024>{{Cite web|title=Better use of vaccines could reduce antibiotic use by 2.5 billion doses annually, says WHO|url=https://www.who.int/news/item/10-10-2024-better-use-of-vaccines-could-reduce-antibiotic-use-by-2.5-billion-doses-annually--says-who|website=[[World Health Organization]]|date=10 October 2024|access-date=11 October 2024}}</ref> Infections from AMR microbes are more challenging to treat and often require costly alternative therapies that may have more severe side effects.<ref name="Saha_2021">{{cite journal | vauthors = Saha M, Sarkar A | title = Review on Multiple Facets of Drug Resistance: A Rising Challenge in the 21st Century | journal = Journal of Xenobiotics | volume = 11 | issue = 4 | pages = 197–214 | date = December 2021 | pmid = 34940513 | pmc = 8708150 | doi = 10.3390/jox11040013 | doi-access = free }}</ref> Preventive measures, such as using narrow-spectrum antibiotics and improving hygiene practices, aim to reduce the spread of resistance.<ref name="Swedish">{{cite book|title=Swedish work on containment of antibiotic resistance – Tools, methods and experiences|publisher=Public Health Agency of Sweden|year=2014|isbn=978-91-7603-011-0|url=http://www.folkhalsomyndigheten.se/pagefiles/17351/Swedish-work-on-containment-of-antibiotic-resistance.pdf|location=Stockholm|pages=16–17, 121–128|access-date=23 July 2015|archive-url=https://web.archive.org/web/20150723081110/http://www.folkhalsomyndigheten.se/pagefiles/17351/Swedish-work-on-containment-of-antibiotic-resistance.pdf|archive-date=23 July 2015|url-status=live}}</ref> |
|||
Certain antibiotic classes are highly associated with colonisation with superbugs compared to other antibiotic classes. The risk for colonisation increases if there is a lack of sensitivity (resistance) of the superbugs to the antibiotic used and high tissue penetration as well as broad spectrum activity against "good bacteria". In the case of [[Methicillin-resistant Staphylococcus aureus|MRSA]], increased rates of MRSA infections are seen with [[glycopeptides]], [[cephalosporin]]s and especially [[quinolones]].<ref>{{cite journal |author=Tacconelli E, De Angelis G, Cataldo MA, Pozzi E, Cauda R |title=Does antibiotic exposure increase the risk of methicillin-resistant Staphylococcus aureus (MRSA) isolation? A systematic review and meta-analysis |journal=J. Antimicrob. Chemother. |volume=61 |issue=1 |pages=26–38 |year=2008 |month=January |pmid=17986491 |doi=10.1093/jac/dkm416 |url=http://jac.oxfordjournals.org/cgi/content/full/61/1/26 }}</ref><ref>{{Cite journal | last1 = Muto | first1 = CA. | last2 = Jernigan | first2 = JA. | last3 = Ostrowsky | first3 = BE. | last4 = Richet | first4 = HM. | last5 = Jarvis | first5 = WR. | last6 = Boyce | first6 = JM. | last7 = Farr | first7 = BM. | title = SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and enterococcus. | journal = Infect Control Hosp Epidemiol | volume = 24 | issue = 5 | pages = 362–86 | month = May | year = 2003 | doi = 10.1086/502213 | pmid = 12785411 }}</ref> In the case of colonisation with [[C difficile]] the high risk antibiotics include cephalosporins and in particular quinolones and [[clindamycin]].<ref>{{cite web |author=Dr Ralf-Peter Vonberg |title=Clostridium difficile: a challenge for hospitals |url=http://www.ihe-online.com/feature-articles/clostridium-difficile-a-challenge-for-hospitals/trackback/1/index.html |work=European Center for Disease Prevention and Control |publisher=IHE |location=Institute for Medical Microbiology and Hospital Epidemiology |accessdate=27 July 2009}}</ref><ref>{{Cite journal | last1 = Kuijper | first1 = EJ. | last2 = van Dissel | first2 = JT. | last3 = Wilcox | first3 = MH. | title = Clostridium difficile: changing epidemiology and new treatment options. | journal = Curr Opin Infect Dis | volume = 20 | issue = 4 | pages = 376–83 | month = Aug | year = 2007 | doi = 10.1097/QCO.0b013e32818be71d | pmid = 17609596 }}</ref> |
|||
The [[World Health Organization|WHO]] claims that AMR is one of the top global public health and development threats, estimating that bacterial AMR was directly responsible for 1.27 million global deaths in 2019 and contributed to 4.95 million deaths.<ref>{{Cite web |title=Antimicrobial resistance |url=https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance |access-date=2024-11-20 |website=www.who.int |language=en}}</ref> Moreover, the WHO and other international bodies warn that AMR could lead to up to 10 million deaths annually by 2050 unless actions are taken.<ref>{{cite news|vauthors=Chanel S, Doherty B|date=2020-09-10|title='Superbugs' a far greater risk than Covid in Pacific, scientist warns|work=The Guardian|url=https://www.theguardian.com/world/2020/sep/10/superbugs-a-far-greater-risk-than-covid-in-pacific-scientist-warns|access-date=2020-09-14|issn=0261-3077|archive-date=5 December 2022|archive-url=https://web.archive.org/web/20221205165241/https://www.theguardian.com/world/2020/sep/10/superbugs-a-far-greater-risk-than-covid-in-pacific-scientist-warns|url-status=live}}</ref> Global initiatives, such as calls for international AMR treaties, emphasize coordinated efforts to limit misuse, fund research, and provide access to necessary antimicrobials in developing nations. However, the [[COVID-19 pandemic]] redirected resources and scientific attention away from AMR, intensifying the challenge.<ref name="pmid33772597">{{cite journal | vauthors = Rodríguez-Baño J, Rossolini GM, Schultsz C, Tacconelli E, Murthy S, Ohmagari N, Holmes A, Bachmann T, Goossens H, Canton R, Roberts AP, Henriques-Normark B, Clancy CJ, Huttner B, Fagerstedt P, Lahiri S, Kaushic C, Hoffman SJ, Warren M, Zoubiane G, Essack S, Laxminarayan R, Plant L | title = Key considerations on the potential impacts of the COVID-19 pandemic on antimicrobial resistance research and surveillance | journal = Trans R Soc Trop Med Hyg | volume = 115| issue = 10| pages = 1122–1129| date = March 2021 | pmid = 33772597 | pmc = 8083707 | doi = 10.1093/trstmh/trab048 }}</ref> |
|||
===In medicine=== |
|||
{{TOC limit|3}} |
|||
The volume of antibiotic prescribed is the major factor in increasing rates of bacterial resistance rather than compliance with antibiotics.<ref name="Pechère JC 2001 S170–3">{{cite journal |author=Pechère JC |title=Patients' interviews and misuse of antibiotics |journal=Clin. Infect. Dis. |volume=33 Suppl 3 |pages=S170–3 |year=2001 |month=September |pmid=11524715 |doi=10.1086/321844 |url=}}</ref> A single dose of antibiotics leads to a greater risk of resistant organisms to that antibiotic in the person for up to a year.<ref>{{cite web |url=http://www.bmj.com/cgi/content/full/340/may18_2/c2096 |title=Effect of antibiotic prescribing in primary care on antimicrobial resistance in individual patients: systematic review and meta-analysis -- Costelloe et al. 340: c2096 -- BMJ |format= |work= |accessdate=}}</ref> |
|||
==Definition== |
|||
Inappropriate prescribing of antibiotics has been attributed to a number of causes including: people who insist on antibiotics, physicians simply prescribe them as they feel they do not have time to explain why they are not necessary, physicians who do not know when to prescribe antibiotics or else are overly cautious for medical legal reasons.<ref>{{cite journal |author=Arnold SR, Straus SE |title=Interventions to improve antibiotic prescribing practices in ambulatory care |journal=Cochrane Database Syst Rev |issue=4 |pages=CD003539 |year=2005 |pmid=16235325 |doi=10.1002/14651858.CD003539.pub2 |url=}}</ref> A third of people for example believe that antibiotics are effective for the [[common cold]]<ref>{{cite journal |author=McNulty CA, Boyle P, Nichols T, Clappison P, Davey P |title=The public's attitudes to and compliance with antibiotics |journal=J. Antimicrob. Chemother. |volume=60 Suppl 1 |pages=i63–8 |year=2007 |month=August |pmid=17656386 |doi=10.1093/jac/dkm161 |url=}}</ref> and 22% of people do not finish a course of antibiotics primarily due to that fact that they feel better (varying from 10% to 44% depending on the country).<ref>{{cite journal |author=Pechère JC, Hughes D, Kardas P, Cornaglia G |title=Non-compliance with antibiotic therapy for acute community infections: a global survey |journal=Int. J. Antimicrob. Agents |volume=29 |issue=3 |pages=245–53 |year=2007 |month=March |pmid=17229552 |doi=10.1016/j.ijantimicag.2006.09.026 |url=}}</ref> Compliance with once daily antibiotics is better than with twice daily antibiotics.<ref>{{cite journal |author=Kardas P |title=Comparison of patient compliance with once-daily and twice-daily antibiotic regimens in respiratory tract infections: results of a randomized trial |journal=J. Antimicrob. Chemother. |volume=59 |issue=3 |pages=531–6 |year=2007 |month=March |pmid=17289766 |doi=10.1093/jac/dkl528 |url=}}</ref> Sub optimum antibiotic concentrations in critically ill people increase the frequency of antibiotic resistance organisms.<ref>{{cite journal |author=Thomas JK, Forrest A, Bhavnani SM, ''et al.'' |title=Pharmacodynamic evaluation of factors associated with the development of bacterial resistance in acutely ill patients during therapy |journal=Antimicrob. Agents Chemother. |volume=42 |issue=3 |pages=521–7 |year=1998 |month=March |pmid=9517926 |pmc=105492 }}</ref> While taking antibiotics doses less than those recommended may increase rates of resistance, shortening the course of antibiotics may actually decrease rates of resistance.<ref name="Pechère JC 2001 S170–3"/><ref>{{cite journal |author=Li JZ, Winston LG, Moore DH, Bent S |title=Efficacy of short-course antibiotic regimens for community-acquired pneumonia: a meta-analysis |journal=Am. J. Med. |volume=120 |issue=9 |pages=783–90 |year=2007 |month=September |pmid=17765048 |doi=10.1016/j.amjmed.2007.04.023 |url=}}</ref> |
|||
[[File:WhatIsDrugResistance.gif|thumb|300x300px|Diagram showing the difference between non-resistant bacteria and drug resistant bacteria. Non-resistant bacteria multiply, and upon drug treatment, the bacteria die. Drug resistant bacteria multiply as well, but upon drug treatment, the bacteria continue to spread.<ref>{{cite web|title=What is Drug Resistance?|url=https://www.niaid.nih.gov/topics/antimicrobialResistance/Understanding/Pages/drugResistanceDefinition.aspx|website=niaid.nih.gov|access-date=26 July 2015|archive-url=https://web.archive.org/web/20150727153042/http://www.niaid.nih.gov/topics/antimicrobialResistance/Understanding/Pages/drugResistanceDefinition.aspx|archive-date=27 July 2015|url-status=live}}</ref>|alt=Diagram showing difference between non-resistance bacteria and drug resistant bacteria]] |
|||
The [[World Health Organization|WHO]] defines antimicrobial resistance as a microorganism's [[drug resistance|resistance to an antimicrobial drug]] that was once able to treat an infection by that microorganism.<ref name=WHO2014/> A person cannot become resistant to antibiotics. Resistance is a property of the microbe, not a person or other organism infected by a microbe.<ref>{{cite web|url=https://www.cdc.gov/getsmart/antibiotic-use/antibiotic-resistance-faqs.html#antibiotic-resistance-concerns|title=CDC: Get Smart: Know When Antibiotics Work|publisher=Cdc.gov|access-date=12 June 2013|archive-url=https://web.archive.org/web/20150429180658/http://www.cdc.gov/getsmart/antibiotic-use/antibiotic-resistance-faqs.html#antibiotic-resistance-concerns|archive-date=29 April 2015|url-status=live|date=29 May 2018}}</ref> All types of microbes can develop drug resistance. Thus, there are antibiotic, antifungal, antiviral and antiparasitic resistance.<ref name="Tanwar_2014" /><ref name="Saha_2021" /> |
|||
Antibiotic resistance is a subset of antimicrobial resistance. This more specific resistance is linked to bacteria and thus broken down into two further subsets, microbiological and clinical. Microbiological resistance is the most common and occurs from genes, [[Mutation|mutated]] or inherited, that allow the bacteria to resist the mechanism to kill the microbe associated with certain antibiotics. Clinical resistance is shown through the failure of many therapeutic techniques where the bacteria that are normally susceptible to a treatment become resistant after surviving the outcome of the treatment. In both cases of acquired resistance, the bacteria can pass the genetic catalyst for resistance through horizontal gene transfer: conjugation, [[Transduction (genetics)|transduction]], or [[Genetic transformation|transformation]]. This allows the resistance to spread across the same species of pathogen or even similar bacterial pathogens.<ref>{{cite journal| vauthors = MacGowan A, Macnaughton E |date=1 October 2017|title=Antibiotic resistance |journal=Medicine |volume=45 |issue=10 |pages=622–628 |doi=10.1016/j.mpmed.2017.07.006 }}</ref> |
|||
Poor hand hygiene by hospital staff has been associated with the spread of resistant organisms<ref>{{cite journal |author=Girou E, Legrand P, Soing-Altrach S, ''et al.'' |title=Association between hand hygiene compliance and methicillin-resistant Staphylococcus aureus prevalence in a French rehabilitation hospital |journal=Infect Control Hosp Epidemiol |volume=27 |issue=10 |pages=1128–30 |year=2006 |month=October |pmid=17006822 |doi=10.1086/507967 |url=}}</ref> and an increase in hand washing compliance results in decreased rates of these organisms.<ref>{{cite journal |author=Swoboda SM, Earsing K, Strauss K, Lane S, Lipsett PA |title=Electronic monitoring and voice prompts improve hand hygiene and decrease nosocomial infections in an intermediate care unit |journal=Crit. Care Med. |volume=32 |issue=2 |pages=358–63 |year=2004 |month=February |pmid=14758148 |doi=10.1097/01.CCM.0000108866.48795.0F |url=}}</ref> |
|||
== Overview == |
|||
===Role of other animals=== |
|||
Drugs are used in animals that are used as human food, such as cows, pigs, chickens, fish, etc., and these drugs can affect the safety of the meat, milk, and eggs produced from those animals and can be the source of superbugs. For example, farm animals, particularly pigs, are believed to be able to infect people with MRSA.<ref>{{cite journal |title=Drug Resistant Infections: Riding Piggyback |journal=The Economist |date= November 29, 2007 |url=http://www.economist.com/displaystory.cfm?story_id=10205187&fsrc=RSS }}</ref> The resistant bacteria in animals due to antibiotic exposure can be transmitted to humans via three pathways, those being through the consumption of meat, from close or direct contact with animals, or through the environment.<ref>{{cite web |author= Schneider K, Garrett L |title= Non-therapeutic Use of Antibiotics in Animal Agriculture, Corresponding Resistance Rates, and What Can be Done About It |url= http://www.cgdev.org/content/article/detail/1422307/ |date= June 19, 2009 }}</ref> |
|||
WHO report released April 2014 stated, "this serious threat is no longer a prediction for the future, it is happening right now in every region of the world and has the potential to affect anyone, of any age, in any country. Antibiotic resistance—when bacteria change so antibiotics no longer work in people who need them to treat infections—is now a major threat to public health."<ref name="who.int">[https://www.who.int/mediacentre/news/releases/2014/amr-report/en/ "WHO's first global report on antibiotic resistance reveals serious, worldwide threat to public health"] {{Webarchive|url=https://web.archive.org/web/20140502044726/http://www.who.int/mediacentre/news/releases/2014/amr-report/en/ |date=2 May 2014 }} Retrieved 2 May 2014</ref> |
|||
The World Health Organization concluded that antibiotics as growth promoters in animal feeds should be prohibited (in the absence of risk assessments). In 1998, European Union health ministers voted to ban four antibiotics widely used to promote animal growth (despite their scientific panel's recommendations). Regulation banning the use of antibiotics in European feed, with the exception of two antibiotics in poultry feeds, became effective in 2006.<ref>{{cite journal | author = Castanon J.I. | year = 2007 | title = History of the use of antibiotic as growth promoters in European poultry feeds | journal = Poult. Sci. | volume = 86 | issue = 11 | pages = 2466–71 | pmid = 17954599 | doi = 10.3382/ps.2007-00249}}</ref> In Scandinavia, there is evidence that the ban has led to a lower prevalence of antimicrobial resistance in (non-hazardous) animal bacterial populations.<ref>{{cite journal | author = Bengtsson B., Wierup M. | year = 2006 | title = Antimicrobial resistance in Scandinavia after ban of antimicrobial growth promoters | journal = Anim. Biotechnol. | volume = 17 | issue = 2 | pages = 147–56 | pmid = 17127526 | doi = 10.1080/10495390600956920}}</ref> In the USA federal agencies do not collect data on antibiotic use in animals but animal to human spread of drug resistant organisms has been demonstrated in research studies. Antibiotics are still used in U.S. animal feed—along with other ingredients which have safety concerns.<ref name="mathew">{{cite journal |author=Mathew AG, Cissell R, Liamthong S |title=Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production |journal=Foodborne Pathog. Dis. |volume=4 |issue=2 |pages=115–33 |year=2007 |pmid=17600481 |doi=10.1089/fpd.2006.0066 |url=}}</ref><ref>{{cite journal |author=Sapkota AR, Lefferts LY, McKenzie S, Walker P |title=What do we feed to food-production animals? A review of animal feed ingredients and their potential impacts on human health |journal=Environ. Health Perspect. |volume=115 |issue=5 |pages=663–70 |year=2007 |month=May |pmid=17520050 |pmc=1867957 |doi=10.1289/ehp.9760 |url=}}</ref> |
|||
Each year, nearly 5 million deaths are associated with AMR globally.<ref name=WHO10October2024/> In 2019, global deaths attributable to AMR numbered 1.27 million in 2019. That same year, AMR may have contributed to 5<!--~4.95 million (3.62–6.57)--> million deaths and one in five people who died due to AMR were children under five years old.<ref name="Murray_2022" /> |
|||
Growing U.S. consumer concern about using antibiotics in animal feed has led to a niche market of "antibiotic-free" animal products, but this small market is unlikely to change entrenched industry-wide practices.<ref>{{cite journal |author=Baker R |title=Health management with reduced antibiotic use - the U.S. experience |journal=Anim. Biotechnol. |volume=17 |issue=2 |pages=195–205 |year=2006 |pmid=17127530 |doi=10.1080/10495390600962274 |url=}}</ref> |
|||
In 2018, WHO considered antibiotic resistance to be one of the biggest threats to global health, [[food security]] and development.<ref name=":4">{{cite web|url=https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance|title=Antibiotic resistance|website=who.int|access-date=2020-03-16|archive-date=21 May 2021|archive-url=https://web.archive.org/web/20210521035847/https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance|url-status=live}}</ref> Deaths attributable to AMR vary by area: |
|||
In 2001, the Union of Concerned Scientists estimated that greater than 70% of the antibiotics used in the US are given to food animals (e.g. chickens, pigs and cattle) in the absence of disease.<ref>[http://www.ucsusa.org/food_and_environment/antibiotics_and_food/hogging-it-estimates-of-antimicrobial-abuse-in-livestock.html Executive summary from the UCS report "Hogging It: Estimates of Antimicrobial Abuse in Livestock", January 2001]</ref> In 2000 the [[US Food and Drug Administration]] (FDA) announced their intention to revoke approval of [[fluoroquinolone]] use in poultry production because of substantial evidence linking it to the emergence of fluoroquinolone resistant [[campylobacter]] infections in humans. The final decision to ban fluoroquinolones from use in poultry production was not made until five years later because of challenges from the food animal and pharmaceutical industries.<ref name="Nelson-2007">{{Cite journal | last1 = Nelson | first1 = JM. | last2 = Chiller | first2 = TM. | last3 = Powers | first3 = JH. | last4 = Angulo | first4 = FJ. | title = Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story. | journal = Clin Infect Dis | volume = 44 | issue = 7 | pages = 977–80 | month = Apr | year = 2007 | doi = 10.1086/512369 | pmid = 17342653 |url=http://www.journals.uchicago.edu/doi/pdf/10.1086/512369 |format=PDF }}</ref> Today, there are two federal bills (S. 549<ref>[http://www.govtrack.us/congress/bill.xpd?bill=s110-549 US Senate Bill S. 549: Preservation of Antibiotics for Medical Treatment Act of 2007]</ref> and H.R. 962<ref>[http://www.govtrack.us/congress/bill.xpd?bill=h110-962 US House Bill H.R. 962: Preservation of Antibiotics for Medical Treatment Act of 2007]</ref>) aimed at phasing out "non-therapeutic" antibiotics in US food animal production. |
|||
{| class="wikitable" |
|||
|+ |
|||
!Place |
|||
!Deaths per 100,000 attributable to AMR<ref name="Murray_2022" /> |
|||
|- |
|||
|North Africa and Middle East |
|||
|11.2 |
|||
|- |
|||
|Southeast and East Asia, and Oceania |
|||
|11.7 |
|||
|- |
|||
|Latin America and Caribbean |
|||
|14.4 |
|||
|- |
|||
|Central and Eastern Europe and Central Asia |
|||
|17.6 |
|||
|- |
|||
|South Asia |
|||
|21.5 |
|||
|- |
|||
|Sub-Saharan Africa |
|||
|23.7 |
|||
|} |
|||
The [[European Centre for Disease Prevention and Control]] calculated that in 2015 there were 671,689 infections in the EU and [[European Economic Area]] caused by antibiotic-resistant bacteria, resulting in 33,110 deaths. Most were acquired in healthcare settings.<ref>{{cite journal | vauthors = Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, Colomb-Cotinat M, Kretzschmar ME, Devleesschauwer B, Cecchini M, Ouakrim DA, Oliveira TC, Struelens MJ, Suetens C, Monnet DL | title = Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis | journal = The Lancet. Infectious Diseases | volume = 19 | issue = 1 | pages = 56–66 | date = January 2019 | pmid = 30409683 | pmc = 6300481 | doi = 10.1016/S1473-3099(18)30605-4 }}</ref><ref>{{cite web |title=Antibiotic-resistant bacteria responsible for over 33,000 deaths in Europe in 2015, study finds |url=https://pharmaceutical-journal.com/article/news/antibiotic-resistant-bacteria-responsible-for-over-33000-deaths-in-europe-in-2015-study-finds |access-date=2023-03-28 |website=The Pharmaceutical Journal |date=7 November 2018|archive-date=28 March 2023 |archive-url=https://web.archive.org/web/20230328155238/https://pharmaceutical-journal.com/article/news/antibiotic-resistant-bacteria-responsible-for-over-33000-deaths-in-europe-in-2015-study-finds |url-status=live }}</ref> In 2019 there were 133,000 deaths caused by AMR.<ref>{{cite journal | title = The burden of bacterial antimicrobial resistance in the WHO European region in 2019: a cross-country systematic analysis | journal = The Lancet. Public Health | volume = 7 | issue = 11 | pages = e897–e913 | date = November 2022 | pmid = 36244350 | pmc = 9630253 | doi = 10.1016/S2468-2667(22)00225-0 | hdl = 10023/26218 | vauthors = Mestrovic T, Robles Aguilar G, Swetschinski LR, Ikuta KS, Gray AP, Davis Weaver N, Han C, Wool EE, Gershberg Hayoon A, Hay SI, Dolecek C, Sartorius B, Murray CJ, Addo IY, Ahinkorah BO, Ahmed A, Aldeyab MA, Allel K, Ancuceanu R, Anyasodor AE, Ausloos M, Barra F, Bhagavathula AS, Bhandari D, Bhaskar S, Cruz-Martins N, Dastiridou A, Dokova K, Dubljanin E, Durojaiye OC }}</ref> |
|||
== |
== Causes == |
||
AMR is driven largely by the misuse and overuse of antimicrobials.<ref name="WHO10October2024"/> Yet, at the same time, many people around the world do not have access to essential antimicrobials.<ref name=WHO10October2024/> This leads to microbes either evolving a defense against drugs used to treat them, or certain strains of microbes that have a natural resistance to antimicrobials becoming much more prevalent than the ones that are easily defeated with medication.<ref>{{cite web|url=http://cambridgemedicine.org/files/10-7244/cmj-2017-03-001/,%20http://cambridgemedicine.org/78-2/|title=Antimicrobial Resistance " Cambridge Medicine Journal|access-date=2020-02-27}}{{Dead link|date=July 2023 |bot=InternetArchiveBot |fix-attempted=yes }}</ref> While antimicrobial resistance does occur naturally over time, the use of antimicrobial agents in a variety of settings both within the healthcare industry and outside of has led to antimicrobial resistance becoming increasingly more prevalent.<ref name="Holmes_2016">{{cite journal | vauthors = Holmes AH, Moore LS, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, Guerin PJ, Piddock LJ | title = Understanding the mechanisms and drivers of antimicrobial resistance | journal = Lancet | volume = 387 | issue = 10014 | pages = 176–87 | date = January 2016 | pmid = 26603922 | doi = 10.1016/S0140-6736(15)00473-0 | hdl-access = free | hdl = 10044/1/32225 | s2cid = 1944665 | url = http://pure-oai.bham.ac.uk/ws/files/25678970/Understanding_the_Mechanisms_and_Drivers_of_AMR_25_Aug_2015.docx | access-date = 5 December 2021 | archive-date = 14 April 2022 | archive-url = https://web.archive.org/web/20220414151453/http://pure-oai.bham.ac.uk/ws/files/25678970/Understanding_the_Mechanisms_and_Drivers_of_AMR_25_Aug_2015.docx | url-status = live }}</ref> |
|||
[[Image:Antibiotic resistance.svg|thumb|right|Schematic representation of how antibiotic resistance evolves via natural selection. The top section represents a population of bacteria before exposure to an antibiotic. The middle section shows the population directly after exposure, the phase in which selection took place. The last section shows the distribution of resistance in a new generation of bacteria. The legend indicates the resistance levels of individuals.]] |
|||
Although many microbes develop resistance to antibiotics over time though natural mutation, overprescribing and inappropriate prescription of antibiotics have accelerated the problem. It is possible that as many as 1 in 3 prescriptions written for antibiotics are unnecessary.<ref name="CDC_2016">{{cite web |date=2016-01-01 |title=CDC Newsroom |url=https://www.cdc.gov/media/releases/2016/p0503-unnecessary-prescriptions.html |access-date=2023-02-28 |website=CDC|archive-date=9 March 2023 |archive-url=https://web.archive.org/web/20230309053532/https://www.cdc.gov/media/releases/2016/p0503-unnecessary-prescriptions.html |url-status=live }}</ref> Every year, approximately 154 million prescriptions for antibiotics are written. Of these, up to 46 million are unnecessary or inappropriate for the condition that the patient has.<ref name="CDC_2016"/> Microbes may naturally develop resistance through genetic mutations that occur during cell division, and although random mutations are rare, many microbes reproduce frequently and rapidly, increasing the chances of members of the population acquiring a mutation that increases resistance.<ref name="Michael_2014">{{cite journal | vauthors = Michael CA, Dominey-Howes D, Labbate M | title = The antimicrobial resistance crisis: causes, consequences, and management | journal = Frontiers in Public Health | volume = 2 | pages = 145 | date = 2014 | pmid = 25279369 | pmc = 4165128 | doi = 10.3389/fpubh.2014.00145 | doi-access = free }}</ref> Many individuals stop taking antibiotics when they begin to feel better. When this occurs, it is possible that the microbes that are less susceptible to treatment still remain in the body. If these microbes are able to continue to reproduce, this can lead to an infection by bacteria that are less susceptible or even resistant to an antibiotic.<ref name="Michael_2014"/> |
|||
Antibiotic resistance can be a result of [[horizontal gene transfer]],<ref>Ochiai, K., Yamanaka, T Kimura K and Sawada, O (1959) Inheritance of drug resistance (and its transfer) between Shigella strains and Between Shigella and E.coli strains. Hihon Iji Shimpor 1861: 34 (in Japanese) |
|||
</ref> and also of unlinked point mutations in the [[pathogen]] [[genome]] and a rate of about 1 in 10<sup>8</sup> per chromosomal replication. The antibiotic action against the pathogen can be seen as an environmental pressure; those bacteria which have a mutation allowing them to survive will live on to reproduce. They will then pass this trait to their offspring, which will result in a fully resistant colony. |
|||
=== Natural occurrence === |
|||
The four main mechanisms by which microorganisms exhibit resistance to antimicrobials are: |
|||
[[File:Antibiotic Resistance Spread.jpg|thumb|A CDC infographic on how antibiotic resistance (a major type of antimicrobial resistance) happens and spreads]] |
|||
# Drug inactivation or modification: e.g. enzymatic deactivation of [[Penicillin|''Penicillin'' G]] in some penicillin-resistant bacteria through the production of [[Beta-lactamases|β-lactamases]]. |
|||
AMR is a naturally occurring process.<ref name="CDC About Antimicrobial Resistance"/> Antimicrobial resistance can evolve naturally due to continued exposure to antimicrobials. [[Natural selection]] means that organisms that are able to adapt to their environment, survive, and continue to produce offspring.<ref name=":5">{{cite web|url=https://evolution.berkeley.edu/evolibrary/article/evo_25|title=Natural selection|website=evolution.berkeley.edu|access-date=2020-03-10|archive-date=30 October 2019|archive-url=https://web.archive.org/web/20191030201404/https://evolution.berkeley.edu/evolibrary/article/evo_25|url-status=live}}</ref> As a result, the types of microorganisms that are able to survive over time with continued attack by certain antimicrobial agents will naturally become more prevalent in the environment, and those without this resistance will become obsolete.<ref name="Holmes_2016" /> |
|||
# Alteration of target site: e.g. alteration of [[Penicillin binding protein|PBP]]—the binding target site of penicillins—in [[MRSA]] and other penicillin-resistant bacteria. |
|||
# Alteration of metabolic pathway: e.g. some [[sulfa drugs|sulfonamide]]-resistant bacteria do not require [[para-aminobenzoic acid]] (PABA), an important precursor for the synthesis of [[folic acid]] and [[nucleic acids]] in bacteria inhibited by sulfonamides. Instead, like mammalian cells, they turn to utilizing preformed folic acid. |
|||
# Reduced drug accumulation: by decreasing drug [[Semipermeable membrane|permeability]] and/or increasing active [[efflux]] (pumping out) of the drugs across the cell surface.<ref name= "pmid19678712">{{cite journal |author= Li, X, Nikadio H |title = Efflux-mediated drug resistance in bacteria: an update.| journal = Drug | volume = 69 | issue = 12 | pages = 1555–623| year = 2009 | pmid = 19678712 | doi = 10.2165/11317030-000000000-00000| url = | issn = |pmc= 2847397 }}</ref> |
|||
Some contemporary antimicrobial resistances have also evolved naturally before the use of antimicrobials of human clinical uses. For instance, [[methicillin]]-resistance evolved as a pathogen of [[Hedgehog|hedgehogs]], possibly as a [[Coevolution|co-evolutionary]] adaptation of the pathogen to hedgehogs that are infected by a [[dermatophyte]] that naturally produces antibiotics.<ref name=":6">{{cite journal | vauthors = Larsen J, Raisen CL, Ba X, Sadgrove NJ, Padilla-González GF, Simmonds MS, Loncaric I, Kerschner H, Apfalter P, Hartl R, Deplano A, Vandendriessche S, Černá Bolfíková B, Hulva P, Arendrup MC, Hare RK, Barnadas C, Stegger M, Sieber RN, Skov RL, Petersen A, Angen Ø, Rasmussen SL, Espinosa-Gongora C, Aarestrup FM, Lindholm LJ, Nykäsenoja SM, Laurent F, Becker K, Walther B, Kehrenberg C, Cuny C, Layer F, Werner G, Witte W, Stamm I, Moroni P, Jørgensen HJ, de Lencastre H, Cercenado E, García-Garrote F, Börjesson S, Hæggman S, Perreten V, Teale CJ, Waller AS, Pichon B, Curran MD, Ellington MJ, Welch JJ, Peacock SJ, Seilly DJ, Morgan FJ, Parkhill J, Hadjirin NF, Lindsay JA, Holden MT, Edwards GF, Foster G, Paterson GK, Didelot X, Holmes MA, Harrison EM, Larsen AR | title = Emergence of methicillin resistance predates the clinical use of antibiotics | journal = Nature | volume = 602 | issue = 7895 | pages = 135–141 | date = February 2022 | pmid = 34987223 | pmc = 8810379 | doi = 10.1038/s41586-021-04265-w | bibcode = 2022Natur.602..135L }}</ref> Also, many [[Soil microbiology|soil fungi and bacteria]] are natural competitors and the original antibiotic [[penicillin]] discovered by [[Alexander Fleming]] rapidly lost clinical effectiveness in treating humans and, furthermore, none of the other natural penicillins (F, K, N, X, O, U1 or U6) are currently in clinical use.{{citation needed|date=April 2023}} |
|||
There are three known mechanisms of [[fluoroquinolone]] resistance. Some types of [[efflux]] pumps can act to decrease intracellular [[quinolone]] concentration.<ref name="pmid9661020">{{cite journal | author = Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, Tsuchiya T | title = NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli | journal = Antimicrob. Agents Chemother. | volume = 42 | issue = 7 | pages = 1778–82 | year = 1998 | month = July | pmid = 9661020 | pmc = 105682 | doi = | url = | issn = }}</ref> In gram-negative bacteria, plasmid-mediated resistance genes produce proteins that can bind to [[DNA gyrase]], protecting it from the action of quinolones. Finally, mutations at key sites in DNA gyrase or [[Topoisomerase IV]] can decrease their binding affinity to quinolones, decreasing the drug's effectiveness.<ref>{{cite journal |author=Robicsek A, Jacoby GA, Hooper DC |title=The worldwide emergence of plasmid-mediated quinolone resistance |journal=Lancet Infect Dis |volume=6 |issue=10 |pages=629–40 |year=2006 |month=October |pmid=17008172 |doi=10.1016/S1473-3099(06)70599-0 |url=http://linkinghub.elsevier.com/retrieve/pii/S1473-3099(06)70599-0}}</ref> Research has shown that the bacterial protein [[LexA]] may play a key role in the acquisition of bacterial mutations giving resistance to quinolones and rifampicin.<ref name="pmid15869329">{{cite journal |author=Cirz RT, Chin JK, Andes DR, de Crécy-Lagard V, Craig WA, Romesberg FE |title=Inhibition of mutation and combating the evolution of antibiotic resistance |journal=PLoS Biol. |volume=3 |issue=6 |pages=e176 |year=2005 |pmid=15869329 |doi=10.1371/journal.pbio.0030176 |url=http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0030176 |pmc=1088971}}</ref> |
|||
Antimicrobial resistance can be acquired from other microbes through swapping genes in a process termed [[horizontal gene transfer]]. This means that once a gene for resistance to an antibiotic appears in a microbial community, it can then spread to other microbes in the community, potentially moving from a non-disease causing microbe to a disease-causing microbe. This process is heavily driven by the [[natural selection]] processes that happen during antibiotic use or misuse.<ref>{{cite journal | vauthors = Crits-Christoph A, Hallowell HA, Koutouvalis K, Suez J | title = Good microbes, bad genes? The dissemination of antimicrobial resistance in the human microbiome | journal = Gut Microbes | volume = 14 | issue = 1 | pages = 2055944 | date = 2022-12-31 | pmid = 35332832 | pmc = 8959533 | doi = 10.1080/19490976.2022.2055944 }}</ref> |
|||
Antibiotic resistance can also be introduced artificially into a microorganism through laboratory protocols, sometimes used as a [[selectable marker]] to examine the mechanisms of gene transfer or to identify individuals that absorbed a piece of DNA that included the resistance gene and another gene of interest. |
|||
Over time, most of the strains of bacteria and infections present will be the type resistant to the antimicrobial agent being used to treat them, making this agent now ineffective to defeat most microbes. With the increased use of antimicrobial agents, there is a speeding up of this natural process.<ref name="Ferri_2017">{{cite journal | vauthors = Ferri M, Ranucci E, Romagnoli P, Giaccone V | title = Antimicrobial resistance: A global emerging threat to public health systems | journal = Critical Reviews in Food Science and Nutrition | volume = 57 | issue = 13 | pages = 2857–2876 | date = September 2017 | pmid = 26464037 | doi = 10.1080/10408398.2015.1077192 | s2cid = 24549694 }}</ref> |
|||
==Resistant pathogens== |
|||
=== ''Staphylococcus aureus''=== |
|||
{{Main|MRSA}} |
|||
''[[Staphylococcus aureus]]'' (colloquially known as "Staph aureus" or a ''Staph infection'') is one of the major resistant pathogens. Found on the [[mucous membranes]] and the [[human skin]] of around a third of the population, it is extremely adaptable to antibiotic pressure. It was one of the earlier bacteria in which [[penicillin]] resistance was found—in 1947, just four years after the drug started being mass-produced. [[Methicillin]] was then the antibiotic of choice, but has since been replaced by [[oxacillin]] due to significant kidney toxicity. MRSA ([[Methicillin-resistant Staphylococcus aureus|methicillin-resistant ''Staphylococcus aureus'']]) was first detected in Britain in 1961 and is now "quite common" in hospitals. MRSA was responsible for 37% of fatal cases of [[sepsis]] in the [[United Kingdom|UK]] in 1999, up from 4% in 1991. Half of all ''S. aureus'' infections in the [[United States|US]] are resistant to penicillin, methicillin, [[tetracycline]] and [[erythromycin]]. |
|||
=== Self-medication === |
|||
This left [[vancomycin]] as the only effective agent available at the time. However, strains with intermediate (4-8 ug/ml) levels of resistance, termed GISA (glycopeptide intermediate ''Staphylococcus aureus'') or VISA (vancomycin intermediate ''Staphylococcus aureus''), began appearing in the late 1990s. The first identified case was in Japan in 1996, and strains have since been found in hospitals in England, France and the US. The first documented strain with complete (>16 ug/ml) resistance to vancomycin, termed VRSA ([[Vancomycin-resistant Staphylococcus aureus|Vancomycin-resistant ''Staphylococcus aureus'']]) appeared in the United States in 2002.<ref>{{Cite pmid|14563898}}</ref> |
|||
In the vast majority of countries, antibiotics can only be prescribed by a doctor and supplied by a pharmacy.<ref>{{cite web |title=Global Database for Tracking Antimicrobial Resistance (AMR) Country Self- Assessment Survey (TrACSS) |url=http://amrcountryprogress.org/ |access-date=2023-03-28 |website=amrcountryprogress.org|archive-date=28 March 2023 |archive-url=https://web.archive.org/web/20230328115257/http://amrcountryprogress.org/ |url-status=live }}</ref> [[Self-medication]] by consumers is defined as "the taking of medicines on one's own initiative or on another person's suggestion, who is not a certified medical professional", and it has been identified as one of the primary reasons for the evolution of antimicrobial resistance.<ref name="Rather_2017">{{cite journal | vauthors = Rather IA, Kim BC, Bajpai VK, Park YH | title = Self-medication and antibiotic resistance: Crisis, current challenges, and prevention | journal = Saudi Journal of Biological Sciences | volume = 24 | issue = 4 | pages = 808–812 | date = May 2017 | pmid = 28490950 | pmc = 5415144 | doi = 10.1016/j.sjbs.2017.01.004 }}</ref> Self-medication with antibiotics is an unsuitable way of using them but a common practice in resource-constrained countries. The practice exposes individuals to the risk of bacteria that have developed antimicrobial resistance.<ref name=nft1>{{cite journal | vauthors = Torres NF, Chibi B, Middleton LE, Solomon VP, Mashamba-Thompson TP | title = Evidence of factors influencing self-medication with antibiotics in low and middle-income countries: a systematic scoping review | journal = Public Health | volume = 168 | pages = 92–101 | date = March 2019 | pmid = 30716570 | doi = 10.1016/j.puhe.2018.11.018 | s2cid = 73434085 }}</ref> Many people resort to this out of necessity, when access to a physician is unavailable, or when patients have a limited amount of time or money to see a doctor.<ref>{{cite journal | vauthors = Ayukekbong JA, Ntemgwa M, Atabe AN | title = The threat of antimicrobial resistance in developing countries: causes and control strategies | journal = Antimicrobial Resistance and Infection Control | volume = 6 | issue = 1 | pages = 47 | date = 2017-05-15 | pmid = 28515903 | pmc = 5433038 | doi = 10.1186/s13756-017-0208-x | doi-access = free }}</ref> This increased access makes it extremely easy to obtain antimicrobials. An example is India, where in the state of [[Punjab]] 73% of the population resorted to treating their minor health issues and chronic illnesses through self-medication.<ref name="Rather_2017" /> |
|||
Self-medication is higher outside the hospital environment, and this is linked to higher use of antibiotics, with the majority of antibiotics being used in the community rather than hospitals. The prevalence of self-medication in [[Developing country|low- and middle-income countries]] (LMICs) ranges from 8.1% to 93%. Accessibility, affordability, and conditions of health facilities, as well as the health-seeking behavior, are factors that influence self-medication in low- and middle-income countries.<ref name=nft1 /> Two significant issues with self-medication are the lack of knowledge of the public on, firstly, the dangerous effects of certain antimicrobials (for example [[ciprofloxacin]] which can cause [[Tendinopathy|tendonitis]], [[tendon rupture]] and [[aortic dissection]])<ref>{{cite journal | vauthors = Chen C, Patterson B, Simpson R, Li Y, Chen Z, Lv Q, Guo D, Li X, Fu W, Guo B | title = Do fluoroquinolones increase aortic aneurysm or dissection incidence and mortality? A systematic review and meta-analysis | journal = Frontiers in Cardiovascular Medicine | volume = 9 | pages = 949538 | date = 2022-08-09 | pmid = 36017083 | pmc = 9396038 | doi = 10.3389/fcvm.2022.949538 | doi-access = free }}</ref><ref>{{cite journal | vauthors = Shu Y, Zhang Q, He X, Liu Y, Wu P, Chen L | title = Fluoroquinolone-associated suspected tendonitis and tendon rupture: A pharmacovigilance analysis from 2016 to 2021 based on the FAERS database | journal = Frontiers in Pharmacology | volume = 13 | pages = 990241 | date = 2022-09-06 | pmid = 36147351 | pmc = 9486157 | doi = 10.3389/fphar.2022.990241 | doi-access = free }}</ref> and, secondly, broad microbial resistance and when to seek medical care if the infection is not clearing. In order to determine the public's knowledge and preconceived notions on antibiotic resistance, a screening of 3,537 articles published in Europe, Asia, and North America was done. Of the 55,225 total people surveyed in the articles, 70% had heard of antibiotic resistance previously, but 88% of those people thought it referred to some type of physical change in the human body.<ref name="Rather_2017" /> |
|||
A ''new'' class of antibiotics, [[Linezolid|oxazolidinones]], became available in the 1990s, and the first commercially available oxazolidinone, [[linezolid]], is comparable to vancomycin in effectiveness against MRSA. Linezolid-resistance in ''[[Staphylococcus aureus]]'' was reported in 2003.{{Citation needed|date=April 2010}} |
|||
=== Clinical misuse === |
|||
CA-MRSA (Community-acquired MRSA) has now emerged as an epidemic that is responsible for rapidly progressive, fatal diseases including necrotizing pneumonia, severe [[sepsis]] and [[necrotizing fasciitis]].<ref name="pmid17146447">{{cite journal |author=Boyle-Vavra S, Daum RS |title=Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin |journal=Lab. Invest. |volume=87 |issue=1 |pages=3–9 |year=2007 |pmid=17146447 |doi=10.1038/labinvest.3700501}}</ref> Methicillin-resistant ''[[Staphylococcus aureus]]'' ([[MRSA]]) is the most frequently identified antimicrobial drug-resistant pathogen in US hospitals. The [[epidemiology]] of infections caused by MRSA is rapidly changing. In the past 10 years, infections caused by this organism have emerged in the community. The 2 MRSA clones in the United States most closely associated with community outbreaks, [[USA400]] (MW2 strain, ST1 lineage) and [[USA300]], often contain [[Panton-Valentine leukocidin]] (PVL) genes and, more frequently, have been associated with skin and soft tissue infections. Outbreaks of community-associated (CA)-MRSA infections have been reported in correctional facilities, among athletic teams, among military recruits, in newborn nurseries, and among [[men who have sex with men]]. CA-MRSA infections now appear to be endemic in many urban regions and cause most CA-S. aureus infections.<ref name="pmid17479885">{{cite journal |author=Maree CL, Daum RS, Boyle-Vavra S, Matayoshi K, Miller LG |title=Community-associated methicillin-resistant Staphylococcus aureus isolates causing healthcare-associated infections |journal=Emerging Infect. Dis. |volume=13 |issue=2 |pages=236–42 |year=2007 |pmid=17479885 |doi= 10.3201/eid1302.060781|url=http://www.cdc.gov/eid/content/13/2/236.htm?s_cid=eid236_e |pmc=2725868}}</ref> |
|||
{{see also|Antibiotic misuse}} |
|||
Clinical misuse by healthcare professionals is another contributor to increased antimicrobial resistance. Studies done in the US show that the indication for treatment of antibiotics, choice of the agent used, and the duration of therapy was incorrect in up to 50% of the cases studied.<ref name="The antibiotic resistance crisis: p" /> In 2010 and 2011 about a third of antibiotic prescriptions in [[Patient#Outpatients and inpatients|outpatient settings]] in the United States were not necessary.<ref>{{cite journal |vauthors=Fleming-Dutra KE, Hersh AL, Shapiro DJ, Bartoces M, Enns EA, File TM, Finkelstein JA, Gerber JS, Hyun DY, Linder JA, Lynfield R, Margolis DJ, May LS, Merenstein D, Metlay JP, Newland JG, Piccirillo JF, Roberts RM, Sanchez GV, Suda KJ, Thomas A, Woo TM, Zetts RM, Hicks LA |date=May 2016 |title=Prevalence of Inappropriate Antibiotic Prescriptions Among US Ambulatory Care Visits, 2010–2011 |journal=JAMA |volume=315 |issue=17 |pages=1864–73 |doi=10.1001/jama.2016.4151 |pmid=27139059 |doi-access=free}}</ref> Another study in an intensive care unit in a major hospital in France has shown that 30% to 60% of prescribed antibiotics were unnecessary.<ref name="The antibiotic resistance crisis: p">{{cite journal | vauthors = Ventola CL | title = The antibiotic resistance crisis: part 1: causes and threats | journal = P & T | volume = 40 | issue = 4 | pages = 277–83 | date = April 2015 | pmid = 25859123 | pmc = 4378521 }}</ref> These inappropriate uses of antimicrobial agents promote the evolution of antimicrobial resistance by supporting the bacteria in developing genetic alterations that lead to resistance.<ref>{{cite journal | vauthors = Strachan CR, Davies J | title = The Whys and Wherefores of Antibiotic Resistance | journal = Cold Spring Harbor Perspectives in Medicine | volume = 7 | issue = 2 | pages = a025171 | date = February 2017 | pmid = 27793964 | pmc = 5287056 | doi = 10.1101/cshperspect.a025171 }}</ref> |
|||
According to research conducted in the US that aimed to evaluate physicians' attitudes and knowledge on antimicrobial resistance in ambulatory settings, only 63% of those surveyed reported antibiotic resistance as a problem in their local practices, while 23% reported the aggressive prescription of antibiotics as necessary to avoid failing to provide adequate care.<ref>{{cite journal | vauthors = Harris A, Chandramohan S, Awali RA, Grewal M, Tillotson G, Chopra T | title = Physicians' attitude and knowledge regarding antibiotic use and resistance in ambulatory settings | journal = American Journal of Infection Control | volume = 47 | issue = 8 | pages = 864–868 | date = August 2019 | pmid = 30926215 | doi = 10.1016/j.ajic.2019.02.009 | s2cid = 88482220 }}</ref> This demonstrates how a majority of doctors underestimate the impact that their own prescribing habits have on antimicrobial resistance as a whole. It also confirms that some physicians may be overly cautious and prescribe antibiotics for both medical or legal reasons, even when clinical indications for use of these medications are not always confirmed. This can lead to unnecessary antimicrobial use, a pattern which may have worsened during the [[COVID-19]] pandemic.<ref name="Joshi">{{cite journal |vauthors=Joshi MP |title=Don't let Covid boost another killer |journal=Knowable Magazine |date=17 February 2021 |doi=10.1146/knowable-021621-1 |doi-access=free |url=https://knowablemagazine.org/article/health-disease/2021/antibiotic-resistance-covid |access-date=10 August 2022 |archive-date=22 October 2021 |archive-url=https://web.archive.org/web/20211022091845/https://knowablemagazine.org/article/health-disease/2021/antibiotic-resistance-covid |url-status=live }}</ref><ref>{{cite journal | vauthors = Rawson TM, Moore LS, Zhu N, Ranganathan N, Skolimowska K, Gilchrist M, Satta G, Cooke G, Holmes A | title = Bacterial and Fungal Coinfection in Individuals With Coronavirus: A Rapid Review To Support COVID-19 Antimicrobial Prescribing | journal = Clinical Infectious Diseases | volume = 71 | issue = 9 | pages = 2459–2468 | date = December 2020 | pmid = 32358954 | pmc = 7197596 | doi = 10.1093/cid/ciaa530 }}</ref> |
|||
===''Streptococcus'' and ''Enterococcus''=== |
|||
Studies have shown that common misconceptions about the effectiveness and necessity of antibiotics to treat common mild illnesses contribute to their overuse.<ref>{{cite web|url=https://dailytargum.com//article/2021/02/rutgers-study-finds-antibiotic-overuse-is-caused-by-misconceptions-financial|title=Rutgers study finds antibiotic overuse is caused by misconceptions, financial incentives|vauthors=Barnes S|website=The Daily Targum|access-date=16 February 2021|archive-date=6 December 2021|archive-url=https://web.archive.org/web/20211206103329/https://dailytargum.com/article/2021/02/rutgers-study-finds-antibiotic-overuse-is-caused-by-misconceptions-financial|url-status=live}}</ref><ref>{{cite journal | vauthors = Blaser MJ, Melby MK, Lock M, Nichter M | title = Accounting for variation in and overuse of antibiotics among humans | journal = BioEssays | volume = 43 | issue = 2 | pages = e2000163 | date = February 2021 | pmid = 33410142 | doi = 10.1002/bies.202000163 | s2cid = 230811912 }}</ref> |
|||
''[[Streptococcus pyogenes]]'' (Group A Streptococcus: GAS) infections can usually be treated with many different antibiotics. Early treatment may reduce the risk of death from invasive group A streptococcal disease. However, even the best medical care does not prevent death in every case. For those with very severe illness, supportive care in an intensive care unit may be needed. For persons with necrotizing fasciitis, surgery often is needed to remove damaged tissue.<ref>{{cite web |author=Division of Bacterial and Mycotic Diseases |title=Group A Streptococcal (GAS) Disease (strep throat, necrotizing fasciitis, impetigo) -- Frequently Asked Questions |url=http://www.cdc.gov/ncidod/dbmd/diseaseinfo/groupastreptococcal_g.htm |date=2005-10-11 |publisher=Centers for Disease Control and Prevention |accessdate=2007-12-11}}</ref> Strains of ''S. pyogenes'' resistant to [[macrolide]] antibiotics have emerged, however all strains remain uniformly sensitive to [[penicillin]].<ref name="pmid15109426">{{cite journal |author=Albrich WC, Monnet DL, Harbarth S |title=Antibiotic selection pressure and resistance in Streptococcus pneumoniae and Streptococcus pyogenes |journal=Emerging Infect. Dis. |volume=10 |issue=3 |pages=514–7 |year=2004 |pmid=15109426 |doi= |url=http://www.cdc.gov/ncidod/eid/vol10no3/03-0252.htm}}</ref> |
|||
Important to the conversation of antibiotic use is the [[Veterinary medicine|veterinary medical system]]. Veterinary oversight is required by law for all medically important antibiotics.<ref>{{Cite web |title=Antimicrobials {{!}} American Veterinary Medical Association |url=https://www.avma.org/resources-tools/one-health/antimicrobial-use-and-antimicrobial-resistance |access-date=2024-04-24 |website=avma.org|archive-date=24 April 2024 |archive-url=https://web.archive.org/web/20240424183923/https://www.avma.org/resources-tools/one-health/antimicrobial-use-and-antimicrobial-resistance |url-status=live }}</ref> Veterinarians use the [[Pharmacokinetics|Pharmacokinetic]]/pharmacodynamic model (PK/PD) approach to ensuring that the correct dose of the drug is delivered to the correct place at the correct timing.<ref>{{cite journal | vauthors = Caneschi A, Bardhi A, Barbarossa A, Zaghini A | title = The Use of Antibiotics and Antimicrobial Resistance in Veterinary Medicine, a Complex Phenomenon: A Narrative Review | journal = Antibiotics | volume = 12 | issue = 3 | pages = 487 | date = March 2023 | pmid = 36978354 | pmc = 10044628 | doi = 10.3390/antibiotics12030487 | doi-access = free }}</ref> |
|||
Resistance of ''[[Streptococcus pneumoniae]]'' to penicillin and other beta-lactams is increasing worldwide. The major mechanism of resistance involves the introduction of mutations in genes encoding penicillin-binding proteins. Selective pressure is thought to play an important role, and use of beta-lactam antibiotics has been implicated as a risk factor for infection and colonization. Streptococcus pneumoniae is responsible for [[pneumonia]], [[bacteremia]], [[otitis media]], [[meningitis]], [[sinusitis]], [[peritonitis]] and [[arthritis]].<ref name="pmid15109426"/> |
|||
=== Pandemics, disinfectants and healthcare systems === |
|||
Penicillin-resistant [[pneumonia]] caused by ''[[Streptococcus pneumoniae]]'' (commonly known as ''pneumococcus''), was first detected in 1967, as was penicillin-resistant [[gonorrhea]]. Resistance to penicillin substitutes is also known as beyond ''S. aureus''. By 1993 ''[[Escherichia coli]]'' was resistant to five [[quinolones|fluoroquinolone]] variants. ''[[Mycobacterium tuberculosis]]'' is commonly resistant to [[isoniazid]] and [[rifampin]] and sometimes universally resistant to the common treatments. Other pathogens showing some resistance include ''[[Salmonella]]'', ''[[Campylobacter]]'', and ''[[Streptococci]]''.{{Citation needed|date=April 2010}} |
|||
Increased antibiotic use during the early waves of the COVID-19 pandemic may exacerbate this [[List of global issues|global health challenge]]<!--/threat/burden-->.<ref>{{cite news|title=Has COVID-19 made the superbug crisis worse?|url=https://globalnews.ca/news/8602057/has-covid-19-made-the-superbug-crisis-worse/|access-date=12 February 2022|work=Global News|archive-date=12 February 2022|archive-url=https://web.archive.org/web/20220212122151/https://globalnews.ca/news/8602057/has-covid-19-made-the-superbug-crisis-worse/|url-status=live}}</ref><ref>{{cite journal | vauthors = Lucien MA, Canarie MF, Kilgore PE, Jean-Denis G, Fénélon N, Pierre M, Cerpa M, Joseph GA, Maki G, Zervos MJ, Dely P, Boncy J, Sati H, Rio AD, Ramon-Pardo P | title = Antibiotics and antimicrobial resistance in the COVID-19 era: Perspective from resource-limited settings | journal = International Journal of Infectious Diseases | volume = 104 | pages = 250–254 | date = March 2021 | pmid = 33434666 | pmc = 7796801 | doi = 10.1016/j.ijid.2020.12.087 }}</ref> Moreover, pandemic burdens on some healthcare systems may contribute to antibiotic-resistant infections.<ref>{{cite web |title=COVID-19 & Antibiotic Resistance |url=https://www.cdc.gov/drugresistance/covid19.html |website=Centers for Disease Control and Prevention |access-date=21 February 2022|date=18 November 2021 |archive-date=21 February 2022 |archive-url=https://web.archive.org/web/20220221120759/https://www.cdc.gov/drugresistance/covid19.html |url-status=live }}</ref> On the other hand, "increased hand hygiene, decreased international travel, and decreased elective hospital procedures may have reduced AMR pathogen selection and spread in the short term" during the COVID-19 pandemic.<ref>{{cite journal | vauthors = Knight GM, Glover RE, McQuaid CF, Olaru ID, Gallandat K, Leclerc QJ, Fuller NM, Willcocks SJ, Hasan R, van Kleef E, Chandler CI | title = Antimicrobial resistance and COVID-19: Intersections and implications | journal = eLife | volume = 10 | date = February 2021 | pmid = 33588991 | pmc = 7886324 | doi = 10.7554/eLife.64139 | s2cid = 231936902 | doi-access = free }}</ref> The use of [[disinfectant]]s such as alcohol-based hand sanitizers, and antiseptic hand wash may also have the potential to increase antimicrobial resistance.<ref>{{cite journal | vauthors = Lu J, Guo J | title = Disinfection spreads antimicrobial resistance | language = EN | journal = Science | volume = 371 | issue = 6528 | pages = 474 | date = January 2021 | pmid = 33510019 | doi = 10.1126/science.abg4380 | s2cid = 231730007 | bibcode = 2021Sci...371..474L | doi-access = free }}</ref> Extensive use of disinfectants can lead to mutations that induce antimicrobial resistance.<ref>{{cite journal | vauthors = Lobie TA, Roba AA, Booth JA, Kristiansen KI, Aseffa A, Skarstad K, Bjørås M | title = Antimicrobial resistance: A challenge awaiting the post-COVID-19 era | journal = International Journal of Infectious Diseases | volume = 111 | pages = 322–325 | date = October 2021 | pmid = 34508864 | pmc = 8425743 | doi = 10.1016/j.ijid.2021.09.003 | s2cid = 237444117 }}</ref> |
|||
A 2024 [[United Nations]] High-Level Meeting on AMR has pledged to reduce deaths associated with bacterial AMR by 10% over the next six years.<ref name=WHO10October2024/><ref>{{Cite web |date=2024-09-26 |title=World Leaders Approve Milestone Commitment To Reduce Deaths From Antibiotic Resistance By 10% By 2030 - Health Policy Watch |url=https://healthpolicy-watch.news/un-high-level-meeting-approves-milestone-commitment-to-reduce-deaths-from-antibiotic-resistance-10-by-2030/ |access-date=2024-09-28 |language=en-US}}</ref> In their first major declaration on the issue since 2016, global leaders also committed to raising $100 million to update and implement AMR action plans.<ref>{{Cite web |title=UN General Assembly High-Level Meeting on antimicrobial resistance 2024 |url=https://www.who.int/news-room/events/detail/2024/09/26/default-calendar/un-general-assembly-high-level-meeting-on-antimicrobial-resistance-2024 |access-date=2024-09-28 |website=www.who.int |language=en}}</ref> However, the final draft of the declaration omitted an earlier target to reduce antibiotic use in animals by 30% by 2030, due to opposition from meat-producing countries and the farming industry. Critics argue this omission is a major weakness, as livestock accounts for around 73% of global sales of antimicrobial agents, including [[Antibiotic|antibiotics]], [[Antiviral drug|antivirals]], and [[Antiparasitic|antiparasitics]]. |
|||
''[[Enterococcus|Enterococcus faecium]]'' is another superbug found in hospitals. [[Penicillin-Resistant Enterococcus]] was seen in 1983, [[vancomycin-resistant enterococcus]] (VRE) in 1987, and [[Linezolid-Resistant Enterococcus]] (LRE) in the late 1990s.{{Citation needed|date=April 2010}} |
|||
=== Environmental pollution === |
|||
===''Pseudomonas aeruginosa''=== |
|||
Considering the complex interactions between humans, animals and the environment, it is also important to consider the environmental aspects and contributors to antimicrobial resistance.<ref>{{cite journal | vauthors = Musoke D, Namata C, Lubega GB, Niyongabo F, Gonza J, Chidziwisano K, Nalinya S, Nuwematsiko R, Morse T | title = The role of Environmental Health in preventing antimicrobial resistance in low- and middle-income countries | journal = Environmental Health and Preventive Medicine | volume = 26 | issue = 1 | pages = 100 | date = October 2021 | pmid = 34610785 | pmc = 8493696 | doi = 10.1186/s12199-021-01023-2 | doi-access = free | bibcode = 2021EHPM...26..100M }}</ref> Although there are still some knowledge gaps in understanding the mechanisms and transmission pathways,<ref name=":1">{{cite journal | vauthors = Fletcher S | title = Understanding the contribution of environmental factors in the spread of antimicrobial resistance | journal = Environmental Health and Preventive Medicine | volume = 20 | issue = 4 | pages = 243–252 | date = July 2015 | pmid = 25921603 | pmc = 4491066 | doi = 10.1007/s12199-015-0468-0 | bibcode = 2015EHPM...20..243F }}</ref> environmental pollution is considered a significant contributor to antimicrobial resistance.<ref name=":2">{{cite journal | vauthors = Ahmad I, Malak HA, Abulreesh HH | title = Environmental antimicrobial resistance and its drivers: a potential threat to public health | journal = Journal of Global Antimicrobial Resistance | volume = 27 | pages = 101–111 | date = December 2021 | pmid = 34454098 | doi = 10.1016/j.jgar.2021.08.001 | doi-access = free }}</ref> Important contributing factors are through "antibiotic residues", "industrial effluents", " [[Agricultural pollution|agricultural runoffs]]", "heavy metals", "[[Biocide|biocides]] and [[Pesticide|pesticides]]" and "sewage and wastewater" that create reservoirs for resistant genes and bacteria that facilitates the transfer of human pathogens.<ref name=":1" /><ref name=":2" /> Unused or expired antibiotics, if not disposed of properly, can enter water systems and soil.<ref name=":2" /> Discharge from pharmaceutical manufacturing and other industrial companies can also introduce antibiotics and other chemicals into the environment.<ref name=":2" /> These factors allow for creating selective pressure for resistant bacteria.<ref name=":2" /> Antibiotics used in livestock and [[aquaculture]] can contaminate soil and water, which promotes resistance in environmental microbes.<ref name=":1" /> Heavy metals such as [[zinc]], copper and [[Mercury (element)|mercury]], and also biocides and pesticides, can co- select for antibiotic resistance,<ref name=":2" /> enhancing their speed.<ref name=":1" /> Inadequate [[Wastewater treatment|treatment of sewage and wastewater]] allows resistant bacteria and genes to spread through water systems.<ref name=":1" /> |
|||
=== Food production === |
|||
''[[Pseudomonas aeruginosa]]'' is a highly prevalent [[opportunistic pathogen]]. One of the most worrisome characteristics of ''[[P. aeruginosa]]'' consists in its low [[antibiotic]] susceptibility. This low susceptibility is attributable to a concerted action of [[efflux (microbiology)|multidrug efflux pumps]] with chromosomally-encoded antibiotic resistance genes (e.g. ''mexAB-oprM'', ''mexXY'' etc.) and the low permeability of the bacterial cellular envelopes.<ref name="Poole2004">{{Cite pmid|14706082}}</ref> Besides intrinsic resistance, ''P. aeruginosa'' easily develop acquired resistance either by [[mutation]] in chromosomally-encoded genes, or by the horizontal gene transfer of antibiotic resistance determinants. Development of [[multidrug resistance]] by ''P. aeruginosa'' isolates requires several different genetic events that include acquisition of different mutations and/or horizontal transfer of antibiotic resistance genes. Hypermutation favours the selection of mutation-driven antibiotic resistance in ''P. aeruginosa'' strains producing chronic infections, whereas the clustering of several different antibiotic resistance genes in [[integron]]s favours the concerted acquisition of antibiotic resistance determinants. Some recent studies have shown that phenotypic resistance associated to [[biofilm]] formation or to the emergence of small-colony-variants may be important in the response of ''P. aeruginosa'' populations to [[antibiotic]]s treatment.<ref name=Cornelis>{{cite book | author = Cornelis P (editor). | title = Pseudomonas: Genomics and Molecular Biology | edition = 1st | publisher = Caister Academic Press | year = 2008 | url=http://www.horizonpress.com/pseudo | isbn=978-1-904455-19-6 }}</ref> |
|||
=== |
==== Livestock ==== |
||
{{main|Antibiotic use in livestock#Antibiotic resistance}} |
|||
[[File:Ar-infographic-950px.jpg|thumb|A CDC infographic on how antibiotic resistance spreads through farm animals]] |
|||
The antimicrobial resistance crisis also extends to the food industry, specifically with food producing animals. With an ever-increasing human population, there is constant pressure to intensify productivity in many agricultural sectors, including the production of meat as a source of protein.<ref>{{cite journal | vauthors = Monger XC, Gilbert AA, Saucier L, Vincent AT | title = Antibiotic Resistance: From Pig to Meat | journal = Antibiotics | volume = 10 | issue = 10 | pages = 1209 | date = October 2021 | pmid = 34680790 | pmc = 8532907 | doi = 10.3390/antibiotics10101209 | doi-access = free }}</ref> Antibiotics are fed to livestock to act as growth supplements, and a preventive measure to decrease the likelihood of infections.<ref>{{cite web |vauthors=Torrella K |date=2023-01-08 |title=Big Meat just can't quit antibiotics |url=https://www.vox.com/future-perfect/2023/1/8/23542789/big-meat-antibiotics-resistance-fda |access-date=2023-01-23 |website=Vox|archive-date=23 January 2023 |archive-url=https://web.archive.org/web/20230123115850/https://www.vox.com/future-perfect/2023/1/8/23542789/big-meat-antibiotics-resistance-fda |url-status=live }}</ref> |
|||
Farmers typically use antibiotics in animal feed to improve growth rates and prevent infections. However, this is illogical as antibiotics are used to treat infections and not prevent infections. 80% of antibiotic use in the U.S. is for agricultural purposes and about 70% of these are medically important.<ref name="auto">{{cite journal | vauthors = Martin MJ, Thottathil SE, Newman TB | title = Antibiotics Overuse in Animal Agriculture: A Call to Action for Health Care Providers | journal = American Journal of Public Health | volume = 105 | issue = 12 | pages = 2409–2410 | date = December 2015 | pmid = 26469675 | pmc = 4638249 | doi = 10.2105/AJPH.2015.302870 }}</ref> Overusing antibiotics gives the bacteria time to adapt leaving higher doses or even stronger antibiotics needed to combat the infection. Though antibiotics for growth promotion were banned throughout the EU in 2006, 40 countries worldwide still use antibiotics to promote growth.<ref>{{cite web |title=Farm antibiotic use |url=https://www.saveourantibiotics.org/the-issue/antibiotic-overuse-in-livestock-farming/ |website=saveourantibiotics.org|access-date=21 March 2024 |archive-date=3 April 2024 |archive-url=https://web.archive.org/web/20240403061957/https://www.saveourantibiotics.org/the-issue/antibiotic-overuse-in-livestock-farming/ |url-status=live }}</ref> |
|||
''[[Clostridium difficile]]'' is a nosocomial pathogen that causes diarrheal disease in hospitals world wide.<ref>Gerding D.N., Johnson S., Peterson L.R., Mulligan M.E. and Silva J. Jr. (1995). ''Clostridium difficile''-associated diarrhea and colitis. Infect. Control. Hosp. Epidemiol. 16:459-477.</ref><ref name=McDonald_2005>{{cite journal |author=McDonald L |title=''Clostridium difficile'': responding to a new threat from an old enemy |journal=Infect. Control. Hosp. Epidemiol. |volume=26 |issue=8 |pages=672–5 |year=2005 |url= http://www.cdc.gov/ncidod/dhqp/pdf/infDis/Cdiff_ICHE08_05.pdf |format=PDF| pmid=16156321 |doi=10.1086/502600}}</ref> [[Clindamycin]]-resistant ''C. difficile'' was reported as the causative agent of large outbreaks of diarrheal disease in hospitals in New York, Arizona, Florida and Massachusetts between 1989 and 1992.<ref name=Johnson1999>{{cite journal |author=Johnson S., Samore M.H., Farrow K.A |title=Epidemics of diarrhea caused by a clindamycin-resistant strain of ''Clostridium difficile'' in four hospitals |journal=New England Journal of Medicine |volume=341 |issue= 23|pages=1645–1651 |year=1999 |url=http://content.nejm.org/cgi/content/full/341/22/1645 |pmid=16322602 |doi=10.1056/NEJM199911253412203 }}</ref> Geographically dispersed outbreaks of ''C. difficile'' strains resistant to [[fluoroquinolone]] antibiotics, such as [[Cipro]] (ciprofloxacin) and [[Levaquin]] (levofloxacin), were also reported in North America in 2005.<ref name=Loo_2005>{{cite journal |author=Loo V, Poirier L, Miller M |title=A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality |journal=N Engl J Med |volume=353 |issue=23 |pages=2442–9 |year=2005 |pmid=16322602 |doi=10.1056/NEJMoa051639}}</ref> |
|||
This can result in the transfer of resistant bacterial strains into the food that humans eat, causing potentially fatal transfer of disease. While the practice of using antibiotics as growth promoters does result in better yields and [[meat]] products, it is a major issue and needs to be decreased in order to prevent antimicrobial resistance.<ref>{{cite journal | vauthors = Tang KL, Caffrey NP, Nóbrega DB, Cork SC, Ronksley PE, Barkema HW, Polachek AJ, Ganshorn H, Sharma N, Kellner JD, Ghali WA | title = Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis | journal = The Lancet. Planetary Health | volume = 1 | issue = 8 | pages = e316–e327 | date = November 2017 | pmid = 29387833 | pmc = 5785333 | doi = 10.1016/S2542-5196(17)30141-9 }}</ref> Though the evidence linking antimicrobial usage in livestock to antimicrobial resistance is limited, the World Health Organization Advisory Group on Integrated Surveillance of Antimicrobial Resistance strongly recommended the reduction of use of medically important antimicrobials in livestock. Additionally, the Advisory Group stated that such antimicrobials should be expressly prohibited for both growth promotion and disease prevention in food producing animals.<ref name="Innes" /> |
|||
===''Salmonella'' and ''E. coli''=== |
|||
''[[Escherichia coli]]'' and ''[[Salmonella]]'' come directly from contaminated food. Of the meat that is contaminated with ''E. coli'', eighty percent of the bacteria are resistant to one or more drugs made; it causes bladder infections that are resistant to antibiotics (“HSUS Fact Sheet”). ''Salmonella'' was first found in humans in the 1970s and in some cases is resistant to as many as nine different antibiotics (“HSUS Fact Sheet”). When both bacterium are spread, serious health conditions arise. Many people are hospitalized each year after becoming infected, and some die as a result. |
|||
By mapping antimicrobial consumption in livestock globally, it was predicted that in 228 countries there would be a total 67% increase in consumption of antibiotics by livestock by 2030. In some countries such as Brazil, Russia, India, China, and South Africa it is predicted that a 99% increase will occur.<ref name="Ferri_2017" /> Several countries have restricted the use of antibiotics in livestock, including Canada, China, Japan, and the US. These restrictions are sometimes associated with a reduction of the [[prevalence]] of antimicrobial resistance in humans.<ref name="Innes">{{cite journal | vauthors = Innes GK, Randad PR, Korinek A, Davis MF, Price LB, So AD, Heaney CD | title = External Societal Costs of Antimicrobial Resistance in Humans Attributable to Antimicrobial Use in Livestock | journal = Annual Review of Public Health | volume = 41 | issue = 1 | pages = 141–157 | date = April 2020 | pmid = 31910712 | pmc = 7199423 | doi = 10.1146/annurev-publhealth-040218-043954 }}</ref> |
|||
===''Acinetobacter baumannii''=== |
|||
On November 5, 2004, the [[Centers for Disease Control and Prevention]] (CDC) reported an increasing number of ''[[Acinetobacter baumannii]]'' bloodstream infections in patients at military medical facilities in which service members injured in the [[Iraq]]/[[Kuwait]] region during [[Iraq war|Operation Iraqi Freedom]] and in [[Afghanistan]] during [[Operation Enduring Freedom]] were treated. Most of these showed [[multidrug resistance]] (MRAB), with a few isolates resistant to all drugs tested.<ref name="pmid15549020">{{cite journal |author= |title=Acinetobacter baumannii infections among patients at military medical facilities treating injured U.S. service members, 2002-2004 |journal=MMWR Morb. Mortal. Wkly. Rep. |volume=53 |issue=45 |pages=1063–6 |year=2004 |pmid=15549020 |doi= |url=http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5345a1.htm |author1= Centers for Disease Control and Prevention (CDC)}}</ref><ref>[http://www.medscape.com/viewarticle/575837 Medscape abstract on Acinetobacter baumannii: Acinetobacter baumannii: An Emerging Multidrug-resistant Threat.]</ref> |
|||
In the United States the [[Veterinary Feed Directive]] went into practice in 2017 dictating that ''All medically important antibiotics to be used in feed or water for food animal species require a veterinary feed directive (VFD) or a prescription.''<ref>{{Cite web |title=Veterinary feed directive (VFD) basics {{!}} American Veterinary Medical Association |url=https://www.avma.org/resources-tools/one-health/antimicrobial-use-and-antimicrobial-resistance/veterinary-feed-directive-basics |access-date=2024-04-24 |website=avma.org|archive-date=24 April 2024 |archive-url=https://web.archive.org/web/20240424183927/https://www.avma.org/resources-tools/one-health/antimicrobial-use-and-antimicrobial-resistance/veterinary-feed-directive-basics |url-status=live }}</ref> |
|||
==Alternatives== |
|||
===Prevention=== |
|||
==== Pesticides ==== |
|||
Rational use of [[antibiotics]] may reduce the chances of development of opportunistic infection by antibiotic-resistant bacteria due to [[dysbacteriosis]]. In one study the use of fluoroquinolones are clearly associated with ''[[Clostridium difficile]]'' infection, which is a leading cause of [[nosocomial]] [[diarrhea]] in the United States,<ref name="pmid12781017">{{cite journal |author=McCusker ME, Harris AD, Perencevich E, Roghmann MC |title=Fluoroquinolone use and Clostridium difficile-associated diarrhea |journal=Emerging Infect. Dis. |volume=9 |issue=6 |pages=730–3 |year=2003 |pmid=12781017 |doi= |url=http://www.cdc.gov/ncidod/eid/vol9no6/02-0385.htm |doi_brokendate=2008-06-24}}</ref> and a major cause of death, worldwide.<ref name="pmid9866738">{{cite journal |author=Frost F, Craun GF, Calderon RL |title=Increasing hospitalization and death possibly due to Clostridium difficile diarrheal disease |journal=Emerging Infect. Dis. |volume=4 |issue=4 |pages=619–25 |year=1998 |pmid=9866738 |doi= 10.3201/eid0404.980412|url=http://www.cdc.gov/ncidod/eid/vol4no4/frost.htm |pmc=2640242 }}</ref> |
|||
{{main|Pesticide resistance}} |
|||
Most [[pesticide]]s protect crops against insects and plants, but in some cases antimicrobial pesticides are used to protect against various microorganisms such as bacteria, viruses, fungi, algae, and protozoa. The overuse of many pesticides in an effort to have a higher yield of crops has resulted in many of these microbes evolving a tolerance against these antimicrobial agents. Currently there are over 4000 antimicrobial pesticides registered with the US [[United States Environmental Protection Agency|Environmental Protection Agency]] (EPA) and sold to market, showing the widespread use of these agents.<ref>{{cite web|url=https://www.epa.gov/pesticide-registration/what-are-antimicrobial-pesticides|title=What are Antimicrobial Pesticides?|last=US EPA|first=OCSPP|date=2013-03-15|website=US EPA|access-date=2020-02-28|archive-date=27 November 2022|archive-url=https://web.archive.org/web/20221127101423/https://www.epa.gov/pesticide-registration/what-are-antimicrobial-pesticides|url-status=live}}</ref> It is estimated that for every single meal a person consumes, 0.3 g of pesticides is used, as 90% of all pesticide use is in agriculture. A majority of these products are used to help defend against the spread of infectious diseases, and hopefully protect public health. But out of the large amount of pesticides used, it is also estimated that less than 0.1% of those antimicrobial agents, actually reach their targets. That leaves over 99% of all pesticides used available to contaminate other resources.<ref>{{cite journal | vauthors = Ramakrishnan B, Venkateswarlu K, Sethunathan N, Megharaj M | title = Local applications but global implications: Can pesticides drive microorganisms to develop antimicrobial resistance? | journal = The Science of the Total Environment | volume = 654 | pages = 177–189 | date = March 2019 | pmid = 30445319 | doi = 10.1016/j.scitotenv.2018.11.041 | s2cid = 53568193 | bibcode = 2019ScTEn.654..177R }}</ref> In soil, air, and water these antimicrobial agents are able to spread, coming in contact with more microorganisms and leading to these microbes evolving mechanisms to tolerate and further resist pesticides. The use of antifungal [[azole]] pesticides that drive environmental azole resistance have been linked to azole resistance cases in the clinical setting.<ref>{{cite journal | vauthors = Rhodes J, Abdolrasouli A, Dunne K, Sewell TR, Zhang Y, Ballard E, Brackin AP, van Rhijn N, Chown H, Tsitsopoulou A, Posso RB, Chotirmall SH, McElvaney NG, Murphy PG, Talento AF, Renwick J, Dyer PS, Szekely A, Bowyer P, Bromley MJ, Johnson EM, Lewis White P, Warris A, Barton RC, Schelenz S, Rogers TR, Armstrong-James D, Fisher MC | title = Population genomics confirms acquisition of drug-resistant Aspergillus fumigatus infection by humans from the environment | journal = Nature Microbiology | volume = 7 | issue = 5 | pages = 663–674 | date = May 2022 | pmid = 35469019 | pmc = 9064804 | doi = 10.1038/s41564-022-01091-2 }}</ref> The same issues confront the novel antifungal classes (e.g. [[orotomide]]s) which are again being used in both the clinic and agriculture.<ref name="Verweij_2022">{{cite journal | vauthors = Verweij PE, Arendrup MC, Alastruey-Izquierdo A, Gold JA, Lockhart SR, Chiller T, White PL | title = Dual use of antifungals in medicine and agriculture: How do we help prevent resistance developing in human pathogens? | journal = Drug Resistance Updates | volume = 65 | pages = 100885 | date = December 2022 | pmid = 36283187 | doi = 10.1016/j.drup.2022.100885 | pmc = 10693676 | s2cid = 253052170 | doi-access = free }}</ref> |
|||
There is clinical evidence that topical dermatological preparations containing [[tea tree oil]] and [[thyme]] oil may be effective in preventing transmittal of [[CA-MRSA]].<ref>David T. Bearden, George P. Allen, and J. Mark Christensen, "Comparative ''in vitro'' activities of topical wound care products against community-associated methicillin-resistant ''Staphylococcus aureus''," ''The Journal of Antimicrobial Chemotherapy'', June 30, 2008, Vol. 62, Number 4, pp. 769–772. [http://jac.oxfordjournals.org/cgi/content/full/62/4/769?maxtoshow=&HITS=10&hits=10&RESULTFORMAT=&fulltext=staphaseptic&searchid=1&FIRSTINDEX=0&resourcetype=HWCIT]</ref> |
|||
=== Wild birds === |
|||
[[Vaccine]]s do not suffer the problem of resistance because a vaccine enhances the body's natural defenses, while an antibiotic operates separately from the body's normal defenses. Nevertheless, new strains may [[Evolution|evolve]] that escape immunity induced by vaccines; for example an update [[Influenza vaccine]] is needed each year. |
|||
Wildlife, including wild and [[Bird migration|migratory birds]], serve as a reservoir for zoonotic disease and antimicrobial-resistant organisms. Birds are a key link between the transmission of zoonotic diseases to human populations. By the same token, increased contact between wild birds and human populations (including domesticated animals), has increased the amount of anti-microbial resistance (AMR) to the bird population.<ref name=":3" /> The introduction of AMR to wild birds positively correlates with human pollution and increased human contact. Additionally, wild birds can participate in [[horizontal gene transfer]] with bacteria, leading to the transmission of antibiotic-resistant genes (ARG).<ref name=":5" /> |
|||
For simplicity, wild bird populations can be divided into two major categories, wild sedentary birds and wild migrating birds. Wild sedentary bird exposure to AMR is through increased contact with densely populated areas, human waste, domestic animals, and domestic animal/livestock waste. Wild migrating birds interact with sedentary birds in different environments along their migration route. This increases the rate and diversity of AMR across varying ecosystems.<ref name=":3" /> |
|||
While theoretically promising, anti-staphylococcal vaccines have shown limited efficacy, because of immunological variation between ''Staphylococcus'' species, and the limited duration of effectiveness of the antibodies produced. Development and testing of more effective vaccines is under way.{{Citation needed|date=April 2010}} |
|||
Neglect of wildlife in the global discussions surrounding [[health security]] and AMR, creates large barriers to true AMR surveillance. The surveillance of anti-microbial resistant organisms in wild birds is a potential metric for the rate of AMR in the environment. This surveillance also allows for further investigation into the transmission routs between different ecosystems and human populations (including domesticated animals and livestock).<ref name=":3" /> Such information gathered from wild bird biomes, can help identify patterns of diseased transmission and better target interventions. These targeted interventions can inform the use of antimicrobial agents and reduce the persistence of multi-drug resistant organisms.<ref name=":4" /><ref name=":6" /> |
|||
The Australian Commonwealth Scientific and Industrial Research Organization ([[CSIRO]]), realizing the need for the reduction of antibiotic use, has been working on two alternatives. One alternative is to prevent diseases by adding [[cytokine]]s instead of antibiotics to animal feed. {{Citation needed|date=April 2010}} These proteins are made in the animal body "naturally" after a disease and are not antibiotics so they do not contribute to the antibiotic resistance problem. Furthermore, studies on using cytokines have shown that they also enhance the growth of animals like the antibiotics now used, but without the drawbacks of non-therapeutic antibiotic use. Cytokines have the potential to achieve the animal growth rates traditionally sought by the use of antibiotics without the contribution of antibiotic resistance associated with the widespread non-therapeutic uses of antibiotics currently utilized in the food animal production industries. Additionally, CSIRO is working on vaccines for diseases.{{Citation needed|date=April 2010}} |
|||
=== Gene transfer from ancient microorganisms === |
|||
===Phage therapy=== |
|||
{{Main|Pathogenic microorganisms in frozen environments}} |
|||
[[Phage therapy]], an approach that has been extensively researched and utilized as a therapeutic agent for over 60 years, especially in the [[Soviet Union]], is an alternative that might help with the problem of resistance. Phage therapy was widely used in the United States until the discovery of antibiotics, in the early 1940s. Bacteriophages or "phages" are viruses that invade bacterial cells and, in the case of lytic phages, disrupt bacterial metabolism and cause the bacterium to [[lysis|lyse]]. Phage therapy is the therapeutic use of lytic [[bacteriophages]] to treat [[pathogenic]] bacterial infections.<ref>{{cite journal |
|||
[[File:Perron_2015_permafrost_antibiotic_resistances.png|thumb|Ancient bacteria found in the permafrost possess a remarkable range of genes which confer resistance to some of the most common antimicrobial classes (red). However, their capacity to resist is also generally lower than of modern bacteria from the same area (black).<ref name="Perron2015" />]] |
|||
| title=Phages and their application against drug-resistant bacteria |
|||
[[Permafrost]] is a term used to refer to any ground that remained frozen for two years or more, with the oldest known examples continuously frozen for around 700,000 years.<ref name="MIT2022">{{cite web |url=https://climate.mit.edu/explainers/permafrost |title=Permafrost | vauthors = McGee D, Gribkoff E |date=4 August 2022 |website=MIT Climate Portal |access-date=27 September 2023 |archive-date=27 September 2023 |archive-url=https://web.archive.org/web/20230927153347/https://climate.mit.edu/explainers/permafrost |url-status=live }}</ref> In the recent decades, permafrost has been rapidly thawing due to [[climate change]].<ref name="AR6_WG1_Chapter922">Fox-Kemper, B., H.T. Hewitt, C. Xiao, G. Aðalgeirsdóttir, S.S. Drijfhout, T.L. Edwards, N.R. Golledge, M. Hemer, R.E. Kopp, G. Krinner, A. Mix, D. Notz, S. Nowicki, I.S. Nurhati, L. Ruiz, J.-B. Sallée, A.B.A. Slangen, and Y. Yu, 2021: [https://www.ipcc.ch/report/ar6/wg1/downloads/report/IPCC_AR6_WGI_Chapter09.pdf Chapter 9: Ocean, Cryosphere and Sea Level Change] {{Webarchive|url=https://web.archive.org/web/20221024162651/https://www.ipcc.ch/report/ar6/wg1/downloads/report/IPCC_AR6_WGI_Chapter09.pdf |date=24 October 2022 }}. In [https://www.ipcc.ch/report/ar6/wg1/ Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change] [Masson-Delmotte, V., P. Zhai, A. Pirani, S.L. Connors, C. Péan, S. Berger, N. Caud, Y. Chen, L. Goldfarb, M.I. Gomis, M. Huang, K. Leitzell, E. Lonnoy, J.B.R. Matthews, T.K. Maycock, T. Waterfield, O. Yelekçi, R. Yu, and B. Zhou (eds.)]. Cambridge University Press, Cambridge, United Kingdom and New York, NY, USA, pp. 1211–1362, doi:10.1017/9781009157896.011.</ref>{{rp|1237}} The cold preserves any [[organic matter]] inside the permafrost, and it is possible for microorganisms to resume their life functions once it thaws. While some common [[pathogen]]s such as [[influenza]], [[smallpox]] or the bacteria associated with [[pneumonia]] have failed to survive intentional attempts to revive them,<ref name="Doucleff2020">{{cite web |url=https://www.npr.org/sections/goatsandsoda/2020/05/19/857992695/are-there-zombie-viruses-like-the-1918-flu-thawing-in-the-permafrost |title=Are There Zombie Viruses — Like The 1918 Flu — Thawing In The Permafrost? | vauthors = Michaeleen D |website=NPR.org |access-date=4 April 2023|archive-date=24 April 2023 |archive-url=https://web.archive.org/web/20230424072912/https://www.npr.org/sections/goatsandsoda/2020/05/19/857992695/are-there-zombie-viruses-like-the-1918-flu-thawing-in-the-permafrost |url-status=live }}</ref> more cold-adapted microorganisms such as [[anthrax]], or several ancient [[plant]] and [[amoeba]] viruses, have successfully survived prolonged thaw.<ref name="Doucleff2016">{{cite web|url=https://www.npr.org/sections/goatsandsoda/2016/08/03/488400947/anthrax-outbreak-in-russia-thought-to-be-result-of-thawing-permafrost|title=Anthrax Outbreak In Russia Thought To Be Result Of Thawing Permafrost|website=NPR.org |url-status=live|archive-url=https://web.archive.org/web/20160922013246/http://www.npr.org/sections/goatsandsoda/2016/08/03/488400947/anthrax-outbreak-in-russia-thought-to-be-result-of-thawing-permafrost|archive-date=2016-09-22|access-date=2016-09-24}}</ref><ref>{{cite journal | vauthors = Ng TF, Chen LF, Zhou Y, Shapiro B, Stiller M, Heintzman PD, Varsani A, Kondov NO, Wong W, Deng X, Andrews TD, Moorman BJ, Meulendyk T, MacKay G, Gilbertson RL, Delwart E | title = Preservation of viral genomes in 700-y-old caribou feces from a subarctic ice patch | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 111 | issue = 47 | pages = 16842–16847 | date = November 2014 | pmid = 25349412 | pmc = 4250163 | doi = 10.1073/pnas.1410429111 | doi-access = free | bibcode = 2014PNAS..11116842N }}</ref><ref name="Legendre 2015 E5327–E5335">{{cite journal | vauthors = Legendre M, Lartigue A, Bertaux L, Jeudy S, Bartoli J, Lescot M, Alempic JM, Ramus C, Bruley C, Labadie K, Shmakova L, Rivkina E, Couté Y, Abergel C, Claverie JM | title = In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 112 | issue = 38 | pages = E5327–E5335 | date = September 2015 | pmid = 26351664 | pmc = 4586845 | doi = 10.1073/pnas.1510795112 | doi-access = free | bibcode = 2015PNAS..112E5327L | jstor = 26465169 }}</ref><ref name="Alempic2023">{{cite journal | vauthors = Alempic JM, Lartigue A, Goncharov AE, Grosse G, Strauss J, Tikhonov AN, Fedorov AN, Poirot O, Legendre M, Santini S, Abergel C, Claverie JM | title = An Update on Eukaryotic Viruses Revived from Ancient Permafrost | journal = Viruses | volume = 15 | issue = 2 | page = 564 | date = February 2023 | pmid = 36851778 | pmc = 9958942 | doi = 10.3390/v15020564 | doi-access = free }}</ref><ref name="Alund2023">{{cite news |url=https://www.usatoday.com/story/news/health/2023/03/09/zombie-virus-frozen-permafrost-revived-after-50-000-years/11434218002/ |title=Scientists revive 'zombie virus' that was frozen for nearly 50,000 years | vauthors = Alund NN |date=9 March 2023 |website=[[USA Today]] |access-date=2023-04-23 |archive-date=2023-04-24 |archive-url=https://web.archive.org/web/20230424073604/https://www.usatoday.com/story/news/health/2023/03/09/zombie-virus-frozen-permafrost-revived-after-50-000-years/11434218002/ |url-status=live }}</ref> |
|||
| journal=J. Chem. Technol. Biotechnol.) |
|||
| year=2001 |
|||
| volume=76 |
|||
| pages=689–699 |
|||
| author=N Chanishvili, T Chanishvili, M. Tediashvili, P.A. Barrow |
|||
| url=http://cat.inist.fr/?aModele=afficheN&cpsidt=1096871 |
|||
| doi=10.1002/jctb.438 }}</ref><ref>{{cite journal |
|||
| title=The use of a novel biodegradable preparation capable of the sustained release of bacteriophages and ciprofloxacin, in the complex treatment of multidrug-resistant Staphylococcus aureus-infected local radiation injuries caused by exposure to Sr90 |
|||
| author=D. Jikia, N. Chkhaidze, E. Imedashvili, I. Mgaloblishvili, G. Tsitlanadze |
|||
| journal=Clinical & Experimental Dermatology |
|||
| year=2005 |
|||
| volume=30 |
|||
| issue=1 |
|||
| page=23 |
|||
| pmid=15663496 |
|||
| url=http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2230.2004.01600.x?journalCode=ced |
|||
| doi = 10.1111/j.1365-2230.2004.01600.x }}</ref><ref>{{cite journal |
|||
|author=Weber-Dabrowska B, Mulczyk M, Górski A |title=Bacteriophages as an efficient therapy for antibiotic-resistant septicemia in man |journal=Transplant. Proc. |volume=35 |issue=4 |pages=1385–6 |year=2003 |month=June |pmid=12826166 |url=http://linkinghub.elsevier.com/retrieve/pii/S0041134503005256 | doi=10.1016/S0041-1345(03)00525-6 |
|||
}}</ref> |
|||
Some scientists have argued that the inability of known [[disease causative agent|causative agent]]s of [[contagious disease]]s to survive being frozen and thawed makes this threat unlikely. Instead, there have been suggestions that when modern pathogenic bacteria interact with the ancient ones, they may, through [[horizontal gene transfer]], pick up [[genetic sequence]]s which are associated with antimicrobial resistance, exacerbating an already difficult issue.<ref name="Sajjad2020">{{cite journal | vauthors = Sajjad W, Rafiq M, Din G, Hasan F, Iqbal A, Zada S, Ali B, Hayat M, Irfan M, Kang S | title = Resurrection of inactive microbes and resistome present in the natural frozen world: Reality or myth? | journal = The Science of the Total Environment | volume = 735 | pages = 139275 | date = September 2020 | pmid = 32480145 | doi = 10.1016/j.scitotenv.2020.139275 | s2cid = 219169932 | doi-access = | bibcode = 2020ScTEn.73539275S }}</ref> Antibiotics to which permafrost bacteria have displayed at least some resistance include [[chloramphenicol]], [[streptomycin]], [[kanamycin]], [[gentamicin]], [[tetracycline]], [[spectinomycin]] and [[neomycin]].<ref name="Miner2021">{{cite journal | vauthors = Miner KR, D'Andrilli J, Mackelprang R, Edwards A, Malaska MJ, Waldrop MP, Miller CE |date=30 September 2021 |title=Emergent biogeochemical risks from Arctic permafrost degradation |journal=Nature Climate Change |volume=11 |issue=1 |pages=809–819 |doi=10.1038/s41558-021-01162-y |bibcode=2021NatCC..11..809M |s2cid=238234156 }}</ref> However, other studies show that resistance levels in ancient bacteria to modern antibiotics remain lower than in the contemporary bacteria from the [[active layer]] of thawed ground above them,<ref name="Perron2015">{{cite journal | vauthors = Perron GG, Whyte L, Turnbaugh PJ, Goordial J, Hanage WP, Dantas G, Desai MM | title = Functional characterization of bacteria isolated from ancient arctic soil exposes diverse resistance mechanisms to modern antibiotics | journal = PLOS ONE | volume = 10 | issue = 3 | pages = e0069533 | date = 25 March 2015 | pmid = 25807523 | pmc = 4373940 | doi = 10.1371/journal.pone.0069533 | doi-access = free | bibcode = 2015PLoSO..1069533P }}</ref> which may mean that this risk is "no greater" than from any other soil.<ref name="Wu2022">{{cite journal| vauthors = Wu R, Trubl G, Taş N, Jansson JK |date=15 April 2022|title=Permafrost as a potential pathogen reservoir|journal=One Earth |volume=5|issue=4|pages=351–360 |doi=10.1016/j.oneear.2022.03.010 |bibcode=2022OEart...5..351W |s2cid=248208195 |doi-access=free}}</ref> |
|||
Bacteriophage therapy is an important alternative to antibiotics in the current era of multidrug resistant pathogens. A review of studies that dealt with the therapeutic use of phages from 1966–1996 and few latest ongoing phage therapy projects via internet showed: phages were used topically, orally or systemically in Polish and Soviet studies. The success rate found in these studies was 80–95% with few gastrointestinal or allergic side effects. British studies also demonstrated significant efficacy of phages against ''[[Escherichia coli]]'', ''[[Acinetobacter]]'' spp., ''[[Pseudomonas]]'' spp and ''[[Staphylococcus aureus]]''. US studies dealt with improving the bioavailability of phage. Phage therapy may prove as an important alternative to antibiotics for treating multidrug resistant pathogens.<ref>{{cite journal | title= Bacteriophage therapy: an alternative to conventional antibiotics | author=Mathur MD, Vidhani S, Mehndiratta PL.| journal=J Assoc Physicians India | year=2003 | volume=51 | pages=593–6 | doi= 10.1258/095646202760159701 | pmid= 12194741 | issue= 8 }}</ref><ref name=McGrath>{{cite book | author = Mc Grath S and van Sinderen D (editors). | title = Bacteriophage: Genetics and Molecular Biology | edition = 1st | publisher = Caister Academic Press | year = 2007 | url=http://www.horizonpress.com/phage | id = [http://www.horizonpress.com/phage ISBN 978-1-904455-14-1 ]}}</ref> |
|||
== |
==Prevention== |
||
[[File:Antibioticresistance diagram.png|thumb|350px|Mission Critical: Preventing Antibiotic Resistance (CDC report, 2014)|alt=Infographic from CDC report on preventing antibiotic resistance]] |
|||
===New medications=== |
|||
Until recently, [[research and development]] (R&D) efforts have provided new drugs in time to treat bacteria that became resistant to older antibiotics. That is no longer the case.{{Citation needed|date=April 2010}} The potential crisis at hand is the result of a marked decrease in industry R&D, and the increasing prevalence of resistant bacteria. Infectious disease physicians are alarmed by the prospect that effective antibiotics may not be available to treat seriously ill patients in the near future. |
|||
There have been increasing public calls for global collective action to address the threat, including a proposal for an international treaty on antimicrobial resistance. Further detail and attention is still needed in order to recognize and measure trends in resistance on the international level; the idea of a global tracking system has been suggested but implementation has yet to occur. A system of this nature would provide insight to areas of high resistance as well as information necessary for evaluating programs, introducing interventions and other changes made to fight or reverse antibiotic resistance.<ref>{{cite journal | vauthors = Rogers Van Katwyk S, Giubilini A, Kirchhelle C, Weldon I, Harrison M, McLean A, Savulescu J, Hoffman SJ | title = Exploring Models for an International Legal Agreement on the Global Antimicrobial Commons: Lessons from Climate Agreements | journal = Health Care Analysis | volume = 31 | issue = 1 | pages = 25–46 | date = March 2023 | pmid = 31965398 | pmc = 10042908 | doi = 10.1007/s10728-019-00389-3 }}</ref><ref>{{cite journal | vauthors = Wilson LA, Van Katwyk SR, Weldon I, Hoffman SJ | title = A Global Pandemic Treaty Must Address Antimicrobial Resistance | journal = The Journal of Law, Medicine & Ethics | volume = 49 | issue = 4 | pages = 688–691 | date = 2021 | pmid = 35006051 | pmc = 8749967 | doi = 10.1017/jme.2021.94 }}</ref> |
|||
The pipeline of new antibiotics is drying up. {{Citation needed|date=April 2010}}Major [[list of pharmaceutical companies|pharmaceutical companies]] are losing interest in the antibiotics market because these drugs may not be as profitable as drugs that treat chronic (long-term) conditions and lifestyle issues.<ref>{{cite web |title=Bad Bugs, No Drugs Executive Summary |url=http://www.idsociety.org/PrintFriendly.aspx?id=5558 |date= |publisher=Infectious Diseases Society of America |accessdate=2007-12-11}}</ref> |
|||
===Duration of antimicrobials=== |
|||
The resistance problem demands that a renewed effort be made to seek antibacterial agents effective against pathogenic bacteria resistant to current antibiotics. One of the possible strategies towards this objective is the rational localization of [[bioactive]] [[phytochemical]]s.{{Citation needed|date=April 2010}} Plants have an almost limitless ability to synthesize [[aromatic]] substances, most of which are [[phenol]]s or their oxygen-substituted derivatives such as [[tannin]]s. Most are secondary [[metabolite]]s, of which at least 12,000 have been isolated, a number estimated to be less than 10% of the total{{Citation needed|date=February 2007}}. In many cases, these substances serve as plant defense mechanisms against [[predation]] by [[microorganism]]s, [[insect]]s, and [[herbivore]]s. Many of the herbs and spices used by humans to season food yield useful medicinal compounds including those having antibacterial activity.<ref name="pmid15831135">{{cite journal |author=Wallace RJ |title=Antimicrobial properties of plant secondary metabolites |journal=Proc Nutr Soc |volume=63 |issue=4 |pages=621–9 |year=2004 |pmid=15831135 |doi=10.1079/PNS2004393}}</ref><ref name="pmid12872531">{{cite journal |author=Thuille N, Fille M, Nagl M |title=Bactericidal activity of herbal extracts |journal=Int J Hyg Environ Health |volume=206 |issue=3 |pages=217–21 |year=2003 |pmid=12872531 |doi=10.1078/1438-4639-00217}}</ref><ref name="pmid12410554">{{cite journal |author=Singh G, Kapoor IP, Pandey SK, Singh UK, Singh RK |title=Studies on essential oils: part 10; antibacterial activity of volatile oils of some spices |journal=Phytother Res |volume=16 |issue=7 |pages=680–2 |year=2002 |pmid=12410554 |doi=10.1002/ptr.951}}</ref> |
|||
Delaying or minimizing the use of antibiotics for certain conditions may help safely reduce their use.<ref name="Spurling_2023">{{cite journal | vauthors = Spurling GK, Dooley L, Clark J, Askew DA | title = Immediate versus delayed versus no antibiotics for respiratory infections | journal = The Cochrane Database of Systematic Reviews | volume = 2023 | issue = 10 | pages = CD004417 | date = October 2023 | pmid = 37791590 | pmc = 10548498 | doi = 10.1002/14651858.CD004417.pub6 | collaboration = Cochrane Acute Respiratory Infections Group }}</ref> Antimicrobial treatment duration should be based on the infection and other health problems a person may have.<ref name="NPS2013">{{cite web|title=Duration of antibiotic therapy and resistance|url=http://www.nps.org.au/publications/health-professional/health-news-evidence/2013/duration-of-antibiotic-therapy|website=NPS Medicinewise|publisher=National Prescribing Service Limited trading, Australia|access-date=22 July 2015|date=13 June 2013|archive-url=https://web.archive.org/web/20150723074759/http://www.nps.org.au/publications/health-professional/health-news-evidence/2013/duration-of-antibiotic-therapy|archive-date=23 July 2015|url-status=dead}}</ref><!-- "When optimising therapy for an infection consider the person's immune status, the infecting agent and the focus of infection." --> For many infections once a person has improved there is little evidence that stopping treatment causes more resistance.<ref name=NPS2013/><!-- "There does not appear to be strong evidence to support the notion that stopping antibiotics before the end of the recommended treatment contributes to increasing resistance" --> Some, therefore, feel that stopping early may be reasonable in some cases.<ref name=NPS2013/><!-- "Therefore, in selected cases, it may be appropriate to stop antibiotic therapy early." --> Other infections, however, do require long courses regardless of whether a person feels better.<ref name=NPS2013/><!-- "For some infections, such as Staphylococcus aureus bacteraemia, enterococcal endocarditis or tuberculosis, clear evidence favours prolonged treatment to prevent relapse" --> |
|||
Delaying antibiotics for ailments such as a sore throat and otitis media may have no difference in the rate of complications compared with immediate antibiotics, for example.<ref name="Spurling_2023" /> When treating [[Respiratory tract infection|respiratory tract infections]], clinical judgement is required as to the appropriate treatment (delayed or immediate antibiotic use).<ref name="Spurling_2023" /> |
|||
Traditional healers have long used plants to prevent or cure infectious conditions. Many of these plants have been investigated scientifically for antimicrobial activity and a large number of plant products have been shown to inhibit growth of pathogenic bacteria. {{Citation needed|date=April 2010}}A number of these agents appear to have structures and modes of action that are distinct from those of the antibiotics in current use, suggesting that [[cross-resistance]] with agents already in use may be minimal. For example the combination of [[5'-methoxyhydnocarpine]] and [[berberine]] in herbs like [[Hydrastis canadensis]] and [[Berberis vulgaris]] can block the MDR-pumps that cause multidrug resistance. This has been shown for [[Staphylococcus aureus]].<ref name="pmid10677479">{{cite journal |author=Stermitz FR, Lorenz P, Tawara JN, Zenewicz LA, Lewis K |title=Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5'-methoxyhydnocarpin, a multidrug pump inhibitor |journal=Proc. Natl. Acad. Sci. U.S.A. |volume=97 |issue=4 |pages=1433–7 |year=2000 |pmid=10677479 |doi=10.1073/pnas.030540597 |url=http://www.pnas.org/cgi/content/full/97/4/1433 |pmc=26451}}</ref> |
|||
The study, "Shorter and Longer Antibiotic Durations for Respiratory Infections: To Fight Antimicrobial Resistance—A Retrospective Cross-Sectional Study in a Secondary Care Setting in the UK," highlights the urgency of reevaluating antibiotic treatment durations amidst the global challenge of antimicrobial resistance (AMR). It investigates the effectiveness of shorter versus longer antibiotic regimens for respiratory tract infections (RTIs) in a UK secondary care setting, emphasizing the need for evidence-based prescribing practices to optimize patient outcomes and combat AMR.<ref>{{cite journal | vauthors = Abdelsalam Elshenawy R, Umaru N, Aslanpour Z | title = Shorter and Longer Antibiotic Durations for Respiratory Infections: To Fight Antimicrobial Resistance-A Retrospective Cross-Sectional Study in a Secondary Care Setting in the UK | journal = Pharmaceuticals | volume = 17 | issue = 3 | pages = 339 | date = March 2024 | pmid = 38543125 | pmc = 10975983 | doi = 10.3390/ph17030339 | doi-access = free }}</ref> |
|||
[[Archaeocin]]s is the name given to a new class of potentially useful antibiotics that are derived from the [[Archaea]] group of organisms. Eight archaeocins have been partially or fully characterized, but hundreds of archaeocins are believed to exist, especially within the [[Archaea|haloarchaea]]. The prevalence of archaeocins is unknown simply because no one has looked for them. The discovery of new archaeocins hinges on recovery and cultivation of archaeal organisms from the environment. For example, samples from a novel hypersaline field site, Wilson Hot Springs, recovered 350 halophilic organisms; preliminary analysis of 75 isolates showed that 48 were archaeal and 27 were bacterial.<ref name=Shand>{{cite book |chapterurl=http://www.horizonpress.com/arch|author= Shand RF; Leyva KJ |year=2008|chapter=Archaeal Antimicrobials: An Undiscovered Country|title=Archaea: New Models for Prokaryotic Biology|publisher=Caister Academic Press|id=[http://www.horizonpress.com/arch ISBN 978-1-904455-27-1]}}</ref> |
|||
===Monitoring and mapping=== |
|||
In research published on October 17, 2008 in [[Cell (journal)|''Cell'']], a team of scientists pinpointed the place on bacteria where the antibiotic [[myxopyronin]] launches its attack, and why that attack is successful. The myxopyronin binds to and inhibits the crucial bacterial enzyme, [[RNA polymerase]]. The myxopyronin changes the structure of the switch-2 segment of the enzyme, inhibiting its function of reading and transmitting DNA code. This prevents RNA polymerase from delivering genetic information to the [[ribosomes]], causing the bacteria to die.<ref>{{cite journal |author=Mukhopadhyay J, Das K, Ismail S, Koppstein D, Jang M, Hudson B, Sarafianos S, Tuske S, Patel J, Jansen R, Irschik H, Arnold E, Ebright RH.|title=The RNA polymerase “switch region” is a target for inhibitors. |doi=10.1016/j.cell.2008.09.033 |journal=Cell |date=2008-10-17 |volume=135 |pages=295–307 |pmid=18957204 |issue=2 |pmc=2580802}}</ref> |
|||
There are multiple national and international monitoring programs for drug-resistant threats, including [[methicillin-resistant Staphylococcus aureus|methicillin-resistant ''Staphylococcus aureus'']] (MRSA), [[Vancomycin-resistant Staphylococcus aureus|vancomycin-resistant ''S. aureus'']] (VRSA), [[Beta-lactamase|extended spectrum beta-lactamase]] (ESBL) producing [[Enterobacterales]], [[Vancomycin-resistant Enterococcus|vancomycin-resistant ''Enterococcus'']] (VRE), and [[Acinetobacter baumannii|multidrug-resistant ''Acinetobacter baumannii'']] (MRAB).<ref name="CDC2013">{{cite web|url=https://www.cdc.gov/drugresistance/biggest_threats.html|title=Biggest Threats – Antibiotic/Antimicrobial Resistance – CDC|website=cdc.gov|access-date=5 May 2016|archive-url=https://web.archive.org/web/20170912115220/https://www.cdc.gov/drugresistance/biggest_threats.html|archive-date=12 September 2017|url-status=live|date=10 September 2018}}</ref> |
|||
ResistanceOpen is an online global map of antimicrobial resistance developed by [[HealthMap]] which displays aggregated data on antimicrobial resistance from publicly available and user submitted data.<ref>{{cite web|url=http://www.healthmap.org/en/index.php|title=HealthMap Resistance|publisher=HealthMap.org Boston Children's Hospital|access-date=15 November 2017|archive-url=https://web.archive.org/web/20171115063743/http://www.healthmap.org/en/index.php|archive-date=15 November 2017|url-status=live}}</ref><ref>{{cite news| vauthors = Scales D |title=Mapping Antibiotic Resistance: Know The Germs in Your Neighborhood|url=http://commonhealth.wbur.org/2015/12/antibiotic-resistance-data|newspaper=Commonhealth|publisher=National Public Radio|access-date=8 December 2015|archive-url=https://web.archive.org/web/20151208101609/http://commonhealth.wbur.org/2015/12/antibiotic-resistance-data|archive-date=8 December 2015|url-status=live}}</ref> The website can display data for a {{convert|25|mi}} radius from a location. Users may submit data from [[antibiogram]]s for individual hospitals or laboratories. European data is from the EARS-Net (European Antimicrobial Resistance Surveillance Network), part of the [[European Centre for Disease Prevention and Control|ECDC]]. ResistanceMap is a website by the [[Center for Disease Dynamics, Economics & Policy]] and provides data on antimicrobial resistance on a global level.<ref>{{cite web|url=https://resistancemap.cddep.org/About.php|title=ResistanceMap|publisher=Center for Disease Dynamics, Economics & Policy|access-date=14 November 2017|archive-url=https://web.archive.org/web/20171114202902/https://resistancemap.cddep.org/About.php|archive-date=14 November 2017|url-status=live}}</ref> |
|||
One of the major causes of antibiotic resistance is the decrease of effective drug concentrations at their target place, due to the increased action of [[ABC transporters]]. Since ABC transporter blockers can be used in combination with current drugs to increase their effective intracellular concentration, the possible impact of ABC transporter inhibitors is of great clinical interest. ABC transporter blockers that may be useful to increase the efficacy of current drugs have entered clinical trials and are available to be used in therapeutic regimes.<ref name= Ponte-SucreA>{{cite book |author= Ponte-Sucre, A (editor)| year=2009 |title=ABC Transporters in Microorganisms | publisher=Caister Academic Press | isbn= 978-1-904455-49-3}}</ref> |
|||
The WHO's AMR global action plan also recommends antimicrobial resistance surveillance in animals.<ref name="WHOAMRactionplan">{{cite web |url= http://www.wpro.who.int/entity/drug_resistance/resources/global_action_plan_eng.pdf |title= Global Action Plan on Antimicrobial Resistance | publisher = WHO |access-date=14 November 2017|archive-url= https://web.archive.org/web/20171031170522/http://www.wpro.who.int/entity/drug_resistance/resources/global_action_plan_eng.pdf |archive-date=31 October 2017|url-status=dead}}</ref> Initial steps in the EU for establishing the veterinary counterpart EARS-Vet (EARS-Net for veterinary medicine) have been made.<ref name="EARS-Vet2021">{{cite journal | vauthors = Mader R, Damborg P, Amat JP, Bengtsson B, Bourély C, Broens EM, Busani L, Crespo-Robledo P, Filippitzi ME, Fitzgerald W, Kaspar H, Madero CM, Norström M, Nykäsenoja S, Pedersen K, Pokludova L, Urdahl AM, Vatopoulos A, Zafeiridis C, Madec JY | title = Building the European Antimicrobial Resistance Surveillance network in veterinary medicine (EARS-Vet) | journal = Euro Surveillance | volume = 26 | issue = 4 | date = January 2021 | pmid = 33509339 | pmc = 7848785 | doi = 10.2807/1560-7917.ES.2021.26.4.2001359 }}</ref> AMR data from pets in particular is scarce, but needed to support antibiotic stewardship in veterinary medicine.<ref name="Feuer2024">{{cite journal | vauthors = Feuer L, Frenzer SK, Merle R, Leistner R, Bäumer W, Bethe A, Lübke-Becker A, Klein B, Bartel A | title = Prevalence of MRSA in canine and feline clinical samples from one-third of veterinary practices in Germany from 2019-2021 | journal = The Journal of Antimicrobial Chemotherapy | volume = 79 | issue = 9 | pages = 2273–2280 | date = September 2024 | pmid = 39007221 | doi = 10.1093/jac/dkae225 }}</ref> |
|||
===Applications=== |
|||
By comparison there is a lack of national and international monitoring programs for antifungal resistance.<ref name="Fisher_2022">{{cite journal | vauthors = Fisher MC, Alastruey-Izquierdo A, Berman J, Bicanic T, Bignell EM, Bowyer P, Bromley M, Brüggemann R, Garber G, Cornely OA, Gurr SJ, Harrison TS, Kuijper E, Rhodes J, Sheppard DC, Warris A, White PL, Xu J, Zwaan B, Verweij PE | title = Tackling the emerging threat of antifungal resistance to human health | journal = Nature Reviews. Microbiology | volume = 20 | issue = 9 | pages = 557–571 | date = September 2022 | pmid = 35352028 | pmc = 8962932 | doi = 10.1038/s41579-022-00720-1 }}</ref> |
|||
Antibiotic resistance is an important tool for [[genetic engineering]]. By constructing a [[plasmid]] which contains an antibiotic resistance gene as well as the gene being engineered or expressed, a researcher can ensure that when bacteria replicate, only the copies which carry along the plasmid survive. This ensures that the gene being manipulated passes along when the bacteria replicates. |
|||
=== Limiting antimicrobial use in humans === |
|||
The most commonly used antibiotics in genetic engineering are generally "older" antibiotics which have largely fallen out of use in clinical practice. These include: |
|||
{{Further|Antibiotic misuse}} |
|||
{{Further|Antimicrobial spectrum}} |
|||
[[Antimicrobial stewardship|Antimicrobial stewardship programmes]] appear useful in reducing rates of antimicrobial resistance.<ref name="pmid28629876">{{cite journal | vauthors = Baur D, Gladstone BP, Burkert F, Carrara E, Foschi F, Döbele S, Tacconelli E | title = Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: a systematic review and meta-analysis | journal = The Lancet. Infectious Diseases | volume = 17 | issue = 9 | pages = 990–1001 | date = September 2017 | pmid = 28629876 | doi = 10.1016/S1473-3099(17)30325-0 }}</ref> The antimicrobial stewardship program will also provide pharmacists with the knowledge to educate patients that antibiotics will not work for a virus for example.<ref>{{cite journal | vauthors = Gallagher JC, Justo JA, Chahine EB, Bookstaver PB, Scheetz M, Suda KJ, Fehrenbacher L, Klinker KP, MacDougall C | title = Preventing the Post-Antibiotic Era by Training Future Pharmacists as Antimicrobial Stewards | journal = American Journal of Pharmaceutical Education | volume = 82 | issue = 6 | pages = 6770 | date = August 2018 | pmid = 30181677 | pmc = 6116871 | doi = 10.5688/ajpe6770 }}</ref> |
|||
* [[ampicillin]] |
|||
* [[kanamycin]] |
|||
* [[tetracycline]] |
|||
* [[chloramphenicol]] |
|||
Excessive antimicrobial use has become one of the top contributors to the evolution of antimicrobial resistance. Since the beginning of the antimicrobial era, antimicrobials have been used to treat a wide range of infectious diseases.<ref name=Andersson2011>{{cite journal | vauthors = Andersson DI, Hughes D | title = Persistence of antibiotic resistance in bacterial populations | journal = FEMS Microbiology Reviews | volume = 35 | issue = 5 | pages = 901–11 | date = September 2011 | pmid = 21707669 | doi = 10.1111/j.1574-6976.2011.00289.x | doi-access = free }}</ref> Overuse of antimicrobials has become the primary cause of rising levels of antimicrobial resistance. The main problem is that doctors are willing to prescribe antimicrobials to ill-informed individuals who believe that antimicrobials can cure nearly all illnesses, including viral infections like the common cold. In an analysis of drug prescriptions, 36% of individuals with a cold or an [[Upper respiratory tract infection|upper respiratory infection]] (both usually viral in origin) were given prescriptions for antibiotics.<ref name=Gilberg2003>{{cite journal | vauthors = Gilberg K, Laouri M, Wade S, Isonaka S | title = Analysis of medication use patterns:apparent overuse of antibiotics and underuse of prescription drugs for asthma, depression, and CHF | journal = Journal of Managed Care Pharmacy | volume = 9 | issue = 3 | pages = 232–7 | year = 2003 | pmid = 14613466 | doi = 10.18553/jmcp.2003.9.3.232 | pmc = 10437266 | s2cid = 25457069 }}</ref> These prescriptions accomplished nothing other than increasing the risk of further evolution of antibiotic resistant bacteria.<ref>{{cite journal | vauthors = Llor C, Bjerrum L | title = Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem | journal = Therapeutic Advances in Drug Safety | volume = 5 | issue = 6 | pages = 229–41 | date = December 2014 | pmid = 25436105 | pmc = 4232501 | doi = 10.1177/2042098614554919 }}</ref> Using antimicrobials without prescription is another driving force leading to the overuse of antibiotics to self-treat diseases like the common cold, cough, fever, and dysentery resulting in an epidemic of antibiotic resistance in countries like Bangladesh, risking its spread around the globe.<ref>{{cite web|date=2021-07-18|title=Pandemic of Antibiotic Resistance Killing Children in Bangladesh|work=Science Trend|url=https://sciencetrend.ca/2021/07/antibiotic-resistance-killing-children/|access-date=2021-08-15|archive-date=29 November 2021|archive-url=https://web.archive.org/web/20211129044831/https://sciencetrend.ca/2021/07/antibiotic-resistance-killing-children/|url-status=dead}}</ref> Introducing strict antibiotic stewardship in the outpatient setting to reduce inappropriate prescribing of antibiotics may reduce the emerging bacterial resistance.<ref>{{cite journal | vauthors = Chisti MJ, Harris JB, Carroll RW, Shahunja KM, Shahid AS, Moschovis PP, Schenkel SR, Hasibur Rahman AS, Shahrin L, Faruk T, Kabir F, Ahmed D, Ahmed T | title = Antibiotic-Resistant Bacteremia in Young Children Hospitalized With Pneumonia in Bangladesh Is Associated With a High Mortality Rate | journal = Open Forum Infectious Diseases | volume = 8 | issue = 7 | pages = ofab260 | date = July 2021 | pmid = 34277885 | pmc = 8280371 | doi = 10.1093/ofid/ofab260 }}</ref> |
|||
Industrially the use of antibiotic resistance is disfavored since maintaining bacterial cultures would require feeding them large quantities of antibiotics. Instead, the use of [[auxotrophic]] bacterial strains (and function-replacement plasmids) is preferred. |
|||
The [[WHO AWaRe]] (Access, Watch, Reserve) guidance and antibiotic book has been introduced to guide antibiotic choice for the 30 most common infections in adults and children to reduce inappropriate prescribing in primary care and hospitals. [[Narrow-spectrum antibiotic]]s are preferred due to their lower resistance potential, and [[broad-spectrum antibiotic]]s are only recommended for people with more severe symptoms. Some antibiotics are more likely to confer resistance, so are kept as reserve antibiotics in the AWaRe book.<ref name="WHO_2022">{{cite book |url=https://www.who.int/publications/i/item/9789240062382 |title=The WHO AWaRe (Access, Watch, Reserve) antibiotic book |publisher=[[World Health Organization]] (WHO) |year=2022 |isbn=978-92-4-006238-2 |location=Geneva |access-date=28 March 2023 |archive-date=13 August 2023 |archive-url=https://web.archive.org/web/20230813134739/https://www.who.int/publications/i/item/9789240062382 |url-status=live }}</ref> |
|||
==See also== |
|||
*[[Antibiotic misuse]] |
|||
*[[Antibiotic tolerance]] |
|||
*[[Drug resistance]] |
|||
*[[Multidrug resistance]] |
|||
*[[Multidrug tolerance]] |
|||
*[[Antibacterial soap]] |
|||
*[[Bacterial conjugation]] |
|||
*[[Drug of last resort]] |
|||
*[[Efflux]] |
|||
*[[Nosocomial infection]] |
|||
*[[Pesticide resistance]] |
|||
*[[Repressor lexA|LexA]] |
|||
*[[Tuberculosis]] |
|||
*[[List of environment topics]] |
|||
*[[Broad-spectrum antibiotic]] |
|||
*[[Center for Disease Dynamics, Economics & Policy]] |
|||
Various diagnostic strategies have been employed to prevent the overuse of antifungal therapy in the clinic, proving a safe alternative to empirical antifungal therapy, and thus underpinning antifungal stewardship schemes.<ref>{{cite journal | vauthors = Talento AF, Qualie M, Cottom L, Backx M, White PL | title = Lessons from an Educational Invasive Fungal Disease Conference on Hospital Antifungal Stewardship Practices across the UK and Ireland | journal = Journal of Fungi | volume = 7 | issue = 10 | pages = 801 | date = September 2021 | pmid = 34682223 | pmc = 8538376 | doi = 10.3390/jof7100801 | doi-access = free }}</ref> |
|||
==References== |
|||
* {{cite journal |author=Soulsby EJ |title=Resistance to antimicrobials in humans and animals |journal=BMJ |volume=331 |issue=7527 |pages=1219–20 |year=2005 |pmid=16308360 |doi=10.1136/bmj.331.7527.1219 |url=http://www.bmj.com/cgi/content/full/331/7527/1219 |pmc=1289307}} |
|||
* {{cite journal |last=Arias |first=Cesar A. |authorlink= |year=2009 |month= |title=Antibiotic-Resistant Bugs in the 21st Century — A Clinical Super-Challenge |journal=[[New England Journal of Medicine]] |volume=360 |issue=5 |pages=439–443 |doi=10.1056/NEJMp0804651 |url= |accessdate= |quote= |pmid=19179312 |last2=Murray |first2=BE }} |
|||
==== At the hospital level ==== |
|||
* “[http://www.csiro.au/solutions/Alternatives-to-antibodies.html Alternatives to Antibiotics Reduce Animal Disease]”. Commonwealth Scientific and Industrial Research Organization. 9 Jan., 2006. 16 Apr., 2009. |
|||
[[Antimicrobial stewardship]] teams in hospitals are encouraging optimal use of antimicrobials.<ref>{{cite journal | vauthors = Doron S, Davidson LE | title = Antimicrobial stewardship | journal = Mayo Clinic Proceedings | volume = 86 | issue = 11 | pages = 1113–23 | date = November 2011 | pmid = 22033257 | pmc = 3203003 | doi = 10.4065/mcp.2011.0358 }}</ref> The goals of antimicrobial stewardship are to help practitioners pick the right drug at the right dose and duration of therapy while preventing misuse and minimizing the development of resistance. Stewardship interventions may reduce the length of stay by an average of slightly over 1 day while not increasing the risk of death.<ref>{{cite journal | vauthors = Davey P, Marwick CA, Scott CL, Charani E, McNeil K, Brown E, Gould IM, Ramsay CR, Michie S | title = Interventions to improve antibiotic prescribing practices for hospital inpatients | journal = The Cochrane Database of Systematic Reviews | volume = 2017 | pages = CD003543 | date = February 2017 | issue = 2 | pmid = 28178770 | pmc = 6464541 | doi = 10.1002/14651858.cd003543.pub4 }}</ref> Dispensing, to discharged in-house patients, the exact number of antibiotic pharmaceutical units necessary to complete an ongoing treatment can reduce antibiotic leftovers within the community as community pharmacies can have antibiotic package inefficiencies.<ref name=":0">{{Cite journal | vauthors = Costa T, Pimentel AC, Mota-Vieira L, Castanha AC |date=2021-05-01 |title=The benefits of a unit dose system in oral antibiotics dispensing: Azorean hospital pharmacists tackling the socioeconomic problem of leftovers in Portugal |url=https://link.springer.com/article/10.1007/s40267-021-00825-2 |journal=Drugs & Therapy Perspectives |language=en |volume=37 |issue=5 |pages=212–221 |doi=10.1007/s40267-021-00825-2 |issn=1179-1977}}</ref> |
|||
==== At the primary care level ==== |
|||
==Footnotes== |
|||
Given the volume of care provided in primary care (general practice), recent strategies have focused on reducing unnecessary antimicrobial prescribing in this setting. Simple interventions, such as written information explaining when taking antibiotics is not necessary, for example in common infections of the upper respiratory tract, have been shown to reduce antibiotic prescribing.<ref>{{cite journal | vauthors = O'Sullivan JW, Harvey RT, Glasziou PP, McCullough A | title = Written information for patients (or parents of child patients) to reduce the use of antibiotics for acute upper respiratory tract infections in primary care | journal = The Cochrane Database of Systematic Reviews | volume = 2016 | pages = CD011360 | date = November 2016 | issue = 11 | pmid = 27886368 | pmc = 6464519 | doi = 10.1002/14651858.CD011360.pub2 }}</ref> Various tools are also available to help professionals decide if prescribing antimicrobials is necessary. |
|||
{{Reflist|2}} |
|||
Parental expectations, driven by the worry for their children's health, can influence how often children are prescribed antibiotics. Parents often rely on their clinician for advice and reassurance. However a lack of plain language information and not having adequate time for consultation negatively impacts this relationship. In effect parents often rely on past experiences in their expectations rather than reassurance from the clinician. Adequate time for consultation and plain language information can help parents make informed decisions and avoid unnecessary antibiotic use.<ref>{{cite journal | vauthors = Bosley H, Henshall C, Appleton JV, Jackson D | title = A systematic review to explore influences on parental attitudes towards antibiotic prescribing in children | journal = Journal of Clinical Nursing | volume = 27 | issue = 5–6 | pages = 892–905 | date = March 2018 | pmid = 28906047 | doi = 10.1111/jocn.14073 | s2cid = 4336064 | url = https://radar.brookes.ac.uk/radar/file/935f9729-d7df-492d-92e9-4afe59a86772/1/fulltext.pdf | access-date = 6 May 2023 | archive-date = 14 October 2023 | archive-url = https://web.archive.org/web/20231014232802/https://radar.brookes.ac.uk/radar/file/935f9729-d7df-492d-92e9-4afe59a86772/1/fulltext.pdf | url-status = live }}</ref> |
|||
==External links== |
|||
* [http://www.who.int/mediacentre/factsheets/fs268/en/index.html WHO fact sheet] |
|||
* [http://www.cdc.gov/ncidod/dhqp/ar_mrsa_spotlight_2006.html CDC Article on Hospital Acquired MRSA] |
|||
* [http://www.cdc.gov/ncidod/dhqp/ar_mrsa_ca.html CDC Article on Community Acquired MRSA] |
|||
* [http://www.cdc.gov/ncidod/dhqp/pdf/ar/mdroGuideline2006.pdf CDC Guideline "Management of Multidrug-Resistant Organisms in Healthcare Settings, 2006"] |
|||
* [http://www.reactgroup.org/ ReAct Action on Antibiotic Resistance] |
|||
<!--* [http://www.cc.nih.gov/hes/vre.html Vancomycin Resistant Enterococcus—Guidelines for Healthcare Workers] (possible deadlink)--> |
|||
* [http://antibiotic.org/ Alliance for the Prudent Use of Antibiotics] |
|||
*{{cite journal |author=Cox LA, Popken DA |title=Quantifying potential human health impacts of animal antibiotic use: enrofloxacin and macrolides in chickens |journal=Risk Anal. |volume=26 |issue=1 |pages=135–46 |year=2006 |month=February |pmid=16492187 |doi=10.1111/j.1539-6924.2006.00723.x |url=}} |
|||
* [http://www.safecarecampaign.org/new_guide/antibiotics/antibiotics_01.html Prudent Use of Antibiotics] |
|||
* [http://www.bacteriophagetherapy.info/ Information about phage therapy – a possible alternative to antibiotics in case of resistant infections] |
|||
* [http://www.gmo-safety.eu/en/gene_transfer/44.docu.html Antibiotic-resistance genes as markers] Once necessary, now undesirable |
|||
* [http://web.archive.org/web/20071105214608/http://cbs5.com/health/local_story_025195614.html CBS Article on Phage Therapy and Antibiotic Resistance] |
|||
* [http://knowledge.allianz.com/en/globalissues/safety_security/health_pandemics/hospital_infections_mrsa.html Hospitals: Breeding the Superbug?] Article on MRSA infections in hospitals, Allianz Knowledge, May 2008 |
|||
* [http://www.eu-burden.info/burden/pages/home.php BURDEN of Resistance and Disease in European Nations - An EU-Project to estimate the financial burden of antibiotic resistance in European Hospitals ] |
|||
* [http://www.extendingthecure.org/ Extending the Cure: Policy Research to Extend Antibiotic Effectiveness ] |
|||
* [http://web.archive.org/web/20071214142631/http://www.fda.gov/oc/antimicrobial/questions.html 2003 New Guidance for Industry on Antimicrobial Drugs for Food Animals Questions and Answers, U.S. FDA] |
|||
*[http://www.scidev.net/en/health/antibiotic-resistance/ SciDev.net Antibiotic Resistance spotlight] The Science and Development Network is an online science and development network focused on news and information important to the developing world |
|||
*[http://www.dobugsneeddrugs.org/ Do Bugs Need Drugs?] |
|||
*[http://www.uu.se/en/node1020 Uppsala university, Resistance to antibiotics – global threat] |
|||
The prescriber should closely adhere to the five rights of drug administration: the right patient, the right drug, the right dose, the right route, and the right time.<ref>{{cite web|url=http://www.ihi.org/resources/pages/improvementstories/fiverightsofmedicationadministration.aspx|title=The Five Rights of Medication Administration|website=ihi.org|date=March 2007 |access-date=30 October 2015|archive-url=https://web.archive.org/web/20151024101457/http://www.ihi.org/resources/pages/improvementstories/fiverightsofmedicationadministration.aspx|archive-date=24 October 2015|url-status=live}}</ref> Microbiological samples should be taken for culture and sensitivity testing before treatment when indicated and treatment potentially changed based on the susceptibility report.<ref name="CDC Mission">{{cite web|url=https://www.cdc.gov/Features/AntibioticResistance/index.html|title=CDC Features – Mission Critical: Preventing Antibiotic Resistance|website=cdc.gov|access-date=22 July 2015|archive-url=https://web.archive.org/web/20171108202412/https://www.cdc.gov/features/antibioticresistance/index.html|archive-date=8 November 2017|url-status=live|date=4 April 2018}}</ref><ref>{{cite journal | vauthors = Leekha S, Terrell CL, Edson RS | title = General principles of antimicrobial therapy | journal = Mayo Clinic Proceedings | volume = 86 | issue = 2 | pages = 156–67 | date = February 2011 | pmid = 21282489 | pmc = 3031442 | doi = 10.4065/mcp.2010.0639 | url = http://www.mayoclinicproceedings.org/article/S0025-6196(11)60140-7/pdf | access-date = 22 July 2015 | archive-date = 15 September 2019 | archive-url = https://web.archive.org/web/20190915154126/https://www.mayoclinicproceedings.org/article/S0025-6196(11)60140-7/pdf | url-status = live }}</ref> |
|||
Health workers and pharmacists can help tackle antibiotic resistance by: enhancing infection prevention and control; only prescribing and dispensing antibiotics when they are truly needed; prescribing and dispensing the right antibiotic(s) to treat the illness.<ref name="who.int" /> A unit dose system implemented in community pharmacies can also reduce antibiotic leftovers at households.<ref name=":0" /> |
|||
==== At the individual level ==== |
|||
People can help tackle resistance by using antibiotics only when infected with a bacterial infection and prescribed by a doctor; completing the full prescription even if the user is feeling better, never sharing antibiotics with others, or using leftover prescriptions.<ref name="who.int" /> Taking antibiotics when not needed won't help the user, but instead give bacteria the option to adapt and leave the user with the side effects that come with the certain type of antibiotic.<ref name="CDC_2022">{{cite web |title=Are you using antibiotics wisely? |url=https://www.cdc.gov/antibiotic-use/do-and-dont.html |website=Centers for Disease Control and Prevention|date=3 January 2022 |access-date=21 March 2024 |archive-date=21 March 2024 |archive-url=https://web.archive.org/web/20240321051000/https://www.cdc.gov/antibiotic-use/do-and-dont.html |url-status=live }}</ref> The CDC recommends that you follow these behaviors so that you avoid these negative side effects and keep the community safe from spreading drug-resistant bacteria.<ref name="CDC_2022"/> Practicing basic bacterial infection prevention courses, such as hygiene, also helps to prevent the spread of antibiotic-resistant bacteria.<ref>{{Cite web|url=https://www.cedars-sinai.org/health-library/articles.html|title=Articles|website=Cedars-Sinai|access-date=23 March 2024|archive-date=30 May 2020|archive-url=https://web.archive.org/web/20200530132825/https://www.cedars-sinai.org/health-library/articles.html|url-status=dead}}</ref> |
|||
====Country examples==== |
|||
* The [[Netherlands]] has the lowest rate of antibiotic prescribing in the [[OECD]], at a rate of 11.4 [[defined daily dose]]s (DDD) per 1,000 people per day in 2011. The defined daily dose (DDD) is a statistical measure of drug consumption, defined by the World Health Organization (WHO).<ref>{{cite web |title=Defined Daily Dose (DDD) |url=https://www.who.int/tools/atc-ddd-toolkit/about-ddd |access-date=2023-03-28 |website=WHO|archive-date=10 May 2023 |archive-url=https://web.archive.org/web/20230510140020/https://www.who.int/tools/atc-ddd-toolkit/about-ddd |url-status=live }}</ref> |
|||
* [[Germany]] and [[Sweden]] also have lower prescribing rates, with Sweden's rate having been declining since 2007. |
|||
* [[Greece]], [[France]] and [[Belgium]] have high prescribing rates for antibiotics of more than 28 DDD.<ref>{{cite web|url=http://www.qualitywatch.org.uk/indicator/antibiotic-prescribing#vis-ref_585|title=Indicator: Antibiotic prescribing|website=QualityWatch|publisher=Nuffield Trust & Health Foundation|access-date=16 July 2015|archive-url=https://web.archive.org/web/20150114213112/http://www.qualitywatch.org.uk/indicator/antibiotic-prescribing#vis-ref_585|archive-date=14 January 2015|url-status=live}}</ref> |
|||
=== Water, sanitation, hygiene === |
|||
Infectious disease control through improved [[WASH|water, sanitation and hygiene (WASH)]] infrastructure needs to be included in the antimicrobial resistance (AMR) agenda. The "Interagency Coordination Group on Antimicrobial Resistance" stated in 2018 that "the spread of pathogens through unsafe water results in a high burden of gastrointestinal disease, increasing even further the need for antibiotic treatment."<ref name="IACG">IACG (2018) [https://www.who.int/antimicrobial-resistance/interagency-coordination-group/IACG_Optimize_use_of_antimicrobials_120718.pdf?ua=1 Reduce unintentional exposure and the need for antimicrobials, and optimize their use IACG Discussion Paper] {{Webarchive|url=https://web.archive.org/web/20210706080228/https://www.who.int/antimicrobial-resistance/interagency-coordination-group/IACG_Optimize_use_of_antimicrobials_120718.pdf?ua=1 |date=6 July 2021 }}, Interagency Coordination Group on Antimicrobial Resistance, [https://web.archive.org/web/20180726093245/http://www.who.int/antimicrobial-resistance/interagency-coordination-group/public-consultation-discussion-papers/en/ public consultation process] at WHO, Geneva, Switzerland</ref> This is particularly a problem in [[developing countries]] where the spread of infectious diseases caused by inadequate WASH standards is a major driver of antibiotic demand.<ref name="Araya">{{cite web|url=https://amr-review.org/sites/default/files/LSE%20AMR%20Capstone.pdf|title=The Impact of Water and Sanitation on Diarrhoeal Disease Burden and Over-Consumption of Anitbiotics.| vauthors = Araya P |date=May 2016|access-date=12 November 2017|archive-url=https://web.archive.org/web/20171001195326/https://amr-review.org/sites/default/files/LSE%20AMR%20Capstone.pdf|archive-date=1 October 2017|url-status=live}}</ref> Growing usage of antibiotics together with persistent infectious disease levels have led to a dangerous cycle in which reliance on antimicrobials increases while the efficacy of drugs diminishes.<ref name="Araya" /> The proper use of infrastructure for water, sanitation and hygiene (WASH) can result in a 47–72 percent decrease of diarrhea cases treated with antibiotics depending on the type of intervention and its effectiveness.<ref name="Araya" /> A reduction of the diarrhea disease burden through improved infrastructure would result in large decreases in the number of diarrhea cases treated with antibiotics. This was estimated as ranging from 5 million in Brazil to up to 590 million in India by the year 2030.<ref name="Araya" /> The strong link between increased consumption and resistance indicates that this will directly mitigate the accelerating spread of AMR.<ref name="Araya" /> Sanitation and water for all by 2030 is [[Sustainable Development Goal 6|Goal Number 6]] of the [[Sustainable Development Goals]].<ref>{{cite web |title=Goal 6: Ensure availability and sustainable management of water and sanitation for all |url=https://sdgs.un.org/goals/goal6 |access-date=2023-04-17 |website=United Nations Department of Economic and Social Affairs |archive-date=24 September 2020 |archive-url=https://archive.today/20200924190731/https://sdgs.un.org/goals/goal6 |url-status=live }}</ref> |
|||
An increase in [[hand washing]] compliance by hospital staff results in decreased rates of resistant organisms.<ref>{{cite journal | vauthors = Swoboda SM, Earsing K, Strauss K, Lane S, Lipsett PA | title = Electronic monitoring and voice prompts improve hand hygiene and decrease nosocomial infections in an intermediate care unit | journal = Critical Care Medicine | volume = 32 | issue = 2 | pages = 358–63 | date = February 2004 | pmid = 14758148 | doi = 10.1097/01.CCM.0000108866.48795.0F | s2cid = 9817602 }}{{subscription required}}</ref> |
|||
Water supply and sanitation infrastructure in health facilities offer significant co-benefits for combatting AMR, and investment should be increased.<ref name="IACG" /> There is much room for improvement: WHO and UNICEF estimated in 2015 that globally 38% of health facilities did not have a source of water, nearly 19% had no toilets and 35% had no water and soap or alcohol-based hand rub for handwashing.<ref>WHO, UNICEF (2015). [https://www.susana.org/en/knowledge-hub/resources-and-publications/library/details/2374 Water, sanitation and hygiene in health care facilities – Status in low and middle income countries and way forward] {{Webarchive|url=https://web.archive.org/web/20180912092005/https://www.susana.org/en/knowledge-hub/resources-and-publications/library/details/2374 |date=12 September 2018 }}. World Health Organization (WHO), Geneva, Switzerland, {{ISBN|978 92 4 150847 6}}</ref> |
|||
=== Industrial wastewater treatment === |
|||
Manufacturers of antimicrobials need to improve the treatment of their wastewater (by using [[industrial wastewater treatment]] processes) to reduce the release of residues into the environment.<ref name="IACG" /> |
|||
=== Limiting antimicrobial use in animals and farming === |
|||
{{Main|Antibiotic use in livestock#Antibiotic resistance}}It is established that the use of [[antibiotics in animal husbandry]] can give rise to AMR resistances in bacteria found in food animals to the antibiotics being administered (through injections or medicated feeds).<ref>{{cite journal |vauthors=Agga GE, Schmidt JW, Arthur TM |date=December 2016 |title=Effects of In-Feed Chlortetracycline Prophylaxis in Beef Cattle on Animal Health and Antimicrobial-Resistant Escherichia coli |journal=Applied and Environmental Microbiology |volume=82 |issue=24 |pages=7197–7204 |bibcode=2016ApEnM..82.7197A |doi=10.1128/AEM.01928-16 |pmc=5118930 |pmid=27736789}}</ref> For this reason only antimicrobials that are deemed "not-clinically relevant" are used in these practices. |
|||
Unlike resistance to antibacterials, antifungal resistance can be driven by [[Arable land|arable farming]], currently there is no regulation on the use of similar antifungal classes in agriculture and the clinic.<ref name="Fisher_2022" /><ref name="Verweij_2022"/> |
|||
Recent studies have shown that the prophylactic use of "non-priority" or "non-clinically relevant" antimicrobials in feeds can potentially, under certain conditions, lead to co-selection of environmental AMR bacteria with resistance to medically important antibiotics.<ref name="Brown EE 2019">{{cite journal |vauthors=Brown EE, Cooper A, Carrillo C, Blais B |date=2019 |title=Selection of Multidrug-Resistant Bacteria in Medicated Animal Feeds |journal=Frontiers in Microbiology|volume=10 |pages=456 |doi=10.3389/fmicb.2019.00456 |pmc=6414793 |pmid=30894847 |doi-access=free}}</ref> The possibility for co-selection of AMR resistances in the food chain pipeline may have far-reaching implications for human health.<ref name="Brown EE 2019" /><ref>{{cite journal |vauthors=Marshall BM, Levy SB |date=October 2011 |title=Food animals and antimicrobials: impacts on human health |journal=Clinical Microbiology Reviews |volume=24 |issue=4 |pages=718–33 |doi=10.1128/CMR.00002-11 |pmc=3194830 |pmid=21976606}}</ref> |
|||
==== Country examples ==== |
|||
===== Europe ===== |
|||
In 1997, European Union health ministers voted to ban [[avoparcin]] and four additional antibiotics used to promote animal growth in 1999.<ref>{{cite journal | vauthors = Casewell M, Friis C, Marco E, McMullin P, Phillips I | title = The European ban on growth-promoting antibiotics and emerging consequences for human and animal health | journal = The Journal of Antimicrobial Chemotherapy | volume = 52 | issue = 2 | pages = 159–61 | date = August 2003 | pmid = 12837737 | doi = 10.1093/jac/dkg313 | doi-access = free }}</ref> In 2006 a ban on the use of antibiotics in European feed, with the exception of two antibiotics in poultry feeds, became effective.<ref>{{cite journal | vauthors = Castanon JI | title = History of the use of antibiotic as growth promoters in European poultry feeds | journal = Poultry Science | volume = 86 | issue = 11 | pages = 2466–71 | date = November 2007 | pmid = 17954599 | doi = 10.3382/ps.2007-00249 | doi-access = free }}{{subscription required}}</ref> In Scandinavia, there is evidence that the ban has led to a lower prevalence of antibiotic resistance in (nonhazardous) animal bacterial populations.<ref>{{cite journal | vauthors = Bengtsson B, Wierup M | title = Antimicrobial resistance in Scandinavia after ban of antimicrobial growth promoters | journal = Animal Biotechnology | volume = 17 | issue = 2 | pages = 147–56 | year = 2006 | pmid = 17127526 | doi = 10.1080/10495390600956920 | s2cid = 34602891 }}{{subscription required}}</ref> As of 2004, several European countries established a decline of antimicrobial resistance in humans through limiting the use of antimicrobials in agriculture and food industries without jeopardizing animal health or economic cost.<ref>{{cite journal | vauthors = Angulo FJ, Baker NL, Olsen SJ, Anderson A, Barrett TJ | title = Antimicrobial use in agriculture: controlling the transfer of antimicrobial resistance to humans | journal = Seminars in Pediatric Infectious Diseases | volume = 15 | issue = 2 | pages = 78–85 | date = April 2004 | pmid = 15185190 | doi = 10.1053/j.spid.2004.01.010 }}</ref> |
|||
===== United States ===== |
|||
The [[United States Department of Agriculture]] (USDA) and the [[Food and Drug Administration]] (FDA) collect data on antibiotic use in humans and in a more limited fashion in animals.<ref name="gao">{{cite web|url=http://www.gao.gov/assets/330/323097.html|title=GAO-11-801, Antibiotic Resistance: Agencies Have Made Limited Progress Addressing Antibiotic Use in Animals|publisher=gao.gov|access-date=25 January 2014|archive-url=https://web.archive.org/web/20131105120254/http://www.gao.gov/assets/330/323097.html|archive-date=5 November 2013|url-status=live}}</ref> About 80% of antibiotic use in the U.S. is for agriculture purposes, and about 70% of these are medically important.<ref name="auto"/> This gives reason for concern about the antibiotic resistance crisis in the U.S. and more reason to monitor it. The FDA first determined in 1977 that there is evidence of emergence of antibiotic-resistant bacterial strains in livestock. The long-established practice of permitting OTC sales of antibiotics (including penicillin and other drugs) to lay animal owners for administration to their own animals nonetheless continued in all states. |
|||
In 2000, the FDA announced their intention to revoke approval of [[fluoroquinolone]] use in poultry production because of substantial evidence linking it to the emergence of fluoroquinolone-resistant ''[[Campylobacter]]'' infections in humans. Legal challenges from the food animal and pharmaceutical industries delayed the final decision to do so until 2006.<ref name="Nelson-2007">{{cite journal | vauthors = Nelson JM, Chiller TM, Powers JH, Angulo FJ | title = Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story | journal = Clinical Infectious Diseases | volume = 44 | issue = 7 | pages = 977–80 | date = April 2007 | pmid = 17342653 | doi = 10.1086/512369 | doi-access = free }}</ref> Fluroquinolones have been banned from extra-label use in food animals in the USA since 2007.<ref>{{cite journal |date=2022-04-29 |title=Extralabel Use and Antimicrobials |url=https://www.fda.gov/animal-veterinary/antimicrobial-resistance/extralabel-use-and-antimicrobials |journal=FDA|access-date=19 April 2023 |archive-date=19 April 2023 |archive-url=https://web.archive.org/web/20230419181246/https://www.fda.gov/animal-veterinary/antimicrobial-resistance/extralabel-use-and-antimicrobials |url-status=live }}</ref> However, they remain widely used in companion and exotic animals.<ref>{{cite journal | vauthors = Pallo-Zimmerman LM, Byron JK, Graves TK | title = Fluoroquinolones: then and now | journal = Compendium | volume = 32 | issue = 7 | pages = E1-9; quiz E9 | date = July 2010 | pmid = 20957609 | url = https://vetfolio-vetstreet.s3.amazonaws.com/1a/a3a710678c11e0a3340050568d17ce/file/PV0710_zimmerman_CE.pdf | access-date = 19 April 2023 | archive-date = 21 June 2023 | archive-url = https://web.archive.org/web/20230621165809/https://vetfolio-vetstreet.s3.amazonaws.com/1a/a3a710678c11e0a3340050568d17ce/file/PV0710_zimmerman_CE.pdf | url-status = live }}</ref> |
|||
===Global action plans and awareness=== |
|||
At the 79th United Nations General Assembly High-Level Meeting on AMR on 26 September 2024, world leaders approved a political declaration committing to a clear set of targets and actions, including reducing the estimated 4.95 million human deaths associated with bacterial AMR annually by 10% by 2030.<ref name=WHO10October2024/> |
|||
The increasing interconnectedness of the world and the fact that new classes of antibiotics have not been developed and approved for more than 25 years highlight the extent to which antimicrobial resistance is a global health challenge.<ref>{{cite web|url=https://www.rand.org/randeurope/research/health/focus-on-antimicrobial-resistance.html|title=RAND Europe Focus on Antimicrobial Resistance (AMR)|website=rand.org|access-date=23 April 2018|archive-url=https://web.archive.org/web/20180421004546/https://www.rand.org/randeurope/research/health/focus-on-antimicrobial-resistance.html|archive-date=21 April 2018|url-status=live}}</ref> A global action plan to tackle the growing problem of resistance to antibiotics and other antimicrobial medicines was endorsed at the Sixty-eighth [[World Health Assembly]] in May 2015.<ref name="WHOAMRactionplan"/> One of the key objectives of the plan is to improve awareness and understanding of antimicrobial resistance through effective communication, education and training. This global action plan developed by the World Health Organization was created to combat the issue of antimicrobial resistance and was guided by the advice of countries and key stakeholders. The WHO's global action plan is composed of five key objectives that can be targeted through different means, and represents countries coming together to solve a major problem that can have future health consequences.<ref name="Ferri_2017" /> These objectives are as follows: |
|||
* improve awareness and understanding of antimicrobial resistance through effective communication, education and training. |
|||
* strengthen the knowledge and evidence base through surveillance and research. |
|||
* reduce the incidence of infection through effective sanitation, hygiene and infection prevention measures. |
|||
* optimize the use of antimicrobial medicines in human and animal health. |
|||
* develop the economic case for sustainable investment that takes account of the needs of all countries and to increase investment in new medicines, diagnostic tools, vaccines and other interventions. |
|||
'''Steps towards progress''' |
|||
* React based in Sweden has produced informative material on AMR for the general public.<ref>{{cite web|url=https://www.reactgroup.org/antibiotic-resistance/the-threat/|title=React|access-date=16 November 2017|archive-url=https://web.archive.org/web/20171116185129/https://www.reactgroup.org/antibiotic-resistance/the-threat/|archive-date=16 November 2017|url-status=live}}</ref> |
|||
* Videos are being produced for the general public to generate interest and awareness.<ref>{{cite web|url=https://www.youtube.com/watch?v=LX6XHvFdzeY| archive-url=https://ghostarchive.org/varchive/youtube/20211111/LX6XHvFdzeY| archive-date=2021-11-11 | url-status=live|title=Antibiotic Resistance: the silent tsunami (youtube video)|date=6 March 2017|website=ReActTube|access-date=17 November 2017}}{{cbignore}}</ref><ref>{{cite web|url=https://www.youtube.com/watch?time_continue=1&v=xZbcwi7SfZE|title=The Antibiotic Apocalypse Explained|date=16 March 2016|website=Kurzgesagt – In a Nutshell|access-date=17 November 2017|archive-date=20 March 2021|archive-url=https://web.archive.org/web/20210320182755/https://www.youtube.com/watch?time_continue=1&v=xZbcwi7SfZE|url-status=live}}</ref> |
|||
* The Irish Department of Health published a National Action Plan on Antimicrobial Resistance in October 2017.<ref>{{cite web|date=October 2017|title=Ireland's National Action Plan on Antimicrobial Resistance 2017 – 2020|url=https://www.lenus.ie/handle/10147/622661|via=Lenus (Irish Health Repository)|access-date=11 January 2019|archive-date=10 August 2022|archive-url=https://web.archive.org/web/20220810172114/https://www.lenus.ie/handle/10147/622661|url-status=live}}</ref> The Strategy for the Control of Antimicrobial Resistance in Ireland (SARI), Iaunched in 2001 developed Guidelines for Antimicrobial Stewardship in Hospitals in Ireland<ref>{{cite book|url=https://www.lenus.ie/handle/10147/303394|title=Guidelines for antimicrobial stewardship in hospitals in Ireland|author=SARI Hospital Antimicrobial Stewardship Working Group|publisher=HSE Health Protection Surveillance Centre (HPSC)|year=2009|isbn=978-0-9551236-7-2|location=Dublin|access-date=11 January 2019|archive-date=5 December 2021|archive-url=https://web.archive.org/web/20211205203416/https://www.lenus.ie/handle/10147/303394|url-status=live}}</ref> in conjunction with the Health Protection Surveillance Centre, these were published in 2009. Following their publication a public information campaign 'Action on Antibiotics<ref>{{cite web|url=https://www.hse.ie/eng/health/hl/hcaiamr/antibiotics/antibiotics.html|archive-url=https://web.archive.org/web/20180528134226/https://www.hse.ie/eng/health/hl/hcaiamr/antibiotics/antibiotics.html|url-status=dead|archive-date=28 May 2018|title=Taking antibiotics for colds and flu? There's no point|website=HSE.ie|access-date=11 January 2019}}</ref>' was launched to highlight the need for a change in antibiotic prescribing. Despite this, antibiotic prescribing remains high with variance in adherence to guidelines.<ref>{{cite journal | vauthors = Murphy M, Bradley CP, Byrne S | title = Antibiotic prescribing in primary care, adherence to guidelines and unnecessary prescribing—an Irish perspective | journal = BMC Family Practice | volume = 13 | pages = 43 | date = May 2012 | issue = 1 | pmid = 22640399 | pmc = 3430589 | doi = 10.1186/1471-2296-13-43 | doi-access = free }}</ref> |
|||
* The United Kingdom published a 20-year vision for antimicrobial resistance that sets out the goal of containing and controlling AMR by 2040.<ref>{{cite web |title=UK 20-year vision for antimicrobial resistance |url=https://www.gov.uk/government/publications/uk-20-year-vision-for-antimicrobial-resistance |access-date=2023-03-28 |website=GOV.UK|archive-date=28 March 2023 |archive-url=https://web.archive.org/web/20230328115249/https://www.gov.uk/government/publications/uk-20-year-vision-for-antimicrobial-resistance |url-status=live }}</ref> The vision is supplemented by a 5-year action plan running from 2019 to 2024, building on the previous action plan (2013–2018).<ref>{{cite web |title=UK 5-year action plan for antimicrobial resistance 2019 to 2024 |url=https://www.gov.uk/government/publications/uk-5-year-action-plan-for-antimicrobial-resistance-2019-to-2024 |access-date=2023-03-28 |website=GOV.UK|archive-date=28 March 2023 |archive-url=https://web.archive.org/web/20230328115251/https://www.gov.uk/government/publications/uk-5-year-action-plan-for-antimicrobial-resistance-2019-to-2024 |url-status=live }}</ref> |
|||
* The World Health Organization has published the 2024 Bacterial Priority Pathogens List which covers 15 families of antibiotic-resistant bacterial pathogens. Notable among these are [[gram-negative bacteria]] resistant to last-resort antibiotics, drug-resistant [[mycobacterium tuberculosis]], and other high-burden resistant pathogens such as [[Salmonella]], [[Shigella]], [[Neisseria gonorrhoeae]], [[Pseudomonas aeruginosa]], and [[Staphylococcus aureus]]. The inclusion of these pathogens in the list underscores their global impact in terms of burden, as well as issues related to transmissibility, treatability, and prevention options. It also reflects the R&D pipeline of new treatments and emerging resistance trends.<ref>{{Cite web |title=WHO bacterial priority pathogens list, 2024: Bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance |url=https://www.who.int/publications-detail-redirect/9789240093461 |access-date=2024-05-20 |website=who.int|archive-date=20 May 2024 |archive-url=https://web.archive.org/web/20240520211731/https://www.who.int/publications-detail-redirect/9789240093461 |url-status=live }}</ref> |
|||
==== Antibiotic Awareness Week ==== |
|||
The World Health Organization has promoted the first World Antibiotic Awareness Week running from 16 to 22 November 2015. The aim of the week is to increase global awareness of antibiotic resistance. It also wants to promote the correct usage of antibiotics across all fields in order to prevent further instances of antibiotic resistance.<ref>{{cite web|url=https://www.who.int/mediacentre/events/2015/world-antibiotic-awareness-week/event/en/|title=World Antibiotic Awareness Week|website=World Health Organization|access-date=20 November 2015|archive-url=https://web.archive.org/web/20151120222039/http://www.who.int/mediacentre/events/2015/world-antibiotic-awareness-week/event/en/|archive-date=20 November 2015|url-status=dead}}</ref> |
|||
World Antibiotic Awareness Week has been held every November since 2015. For 2017, the Food and Agriculture Organization of the United Nations (FAO), the World Health Organization (WHO) and the [[World Organisation for Animal Health]] (OIE) are together calling for responsible use of antibiotics in humans and animals to reduce the emergence of antibiotic resistance.<ref>{{cite web|url=https://www.who.int/campaigns/world-antibiotic-awareness-week/2017/launch-event/en/|title=World Antibiotic Awareness Week|website=WHO|access-date=14 November 2017|archive-url=https://web.archive.org/web/20171113074006/http://www.who.int/campaigns/world-antibiotic-awareness-week/2017/launch-event/en/|archive-date=13 November 2017|url-status=dead}}</ref> |
|||
'''United Nations''' |
|||
In 2016 the Secretary-General of the [[United Nations]] convened the Interagency Coordination Group (IACG) on Antimicrobial Resistance.<ref name="WHO_2">{{cite web|url=https://www.who.int/antimicrobial-resistance/interagency-coordination-group/en/|archive-url=https://web.archive.org/web/20170320110535/http://www.who.int/antimicrobial-resistance/interagency-coordination-group/en/|url-status=dead|archive-date=20 March 2017|title=WHO {{!}} UN Interagency Coordination Group (IACG) on Antimicrobial Resistance|website=WHO|access-date=7 August 2019}}</ref> The IACG worked with international organizations and experts in human, animal, and plant health to create a plan to fight antimicrobial resistance.<ref name="WHO_2" /> Their report released in April 2019 highlights the seriousness of antimicrobial resistance and the threat it poses to world health. It suggests five recommendations for member states to follow in order to tackle this increasing threat. The IACG recommendations are as follows:<ref>{{cite report |url=https://www.who.int/docs/default-source/documents/no-time-to-wait-securing-the-future-from-drug-resistant-infections-en.pdf |title=No Time to Wait: Securing the Future from Drug-Resistant Infections |last=Interagency Coordination Group (IACG) |date=April 2019 |access-date=2023-04-19 |archive-date=19 April 2023 |archive-url=https://web.archive.org/web/20230419181246/https://www.who.int/docs/default-source/documents/no-time-to-wait-securing-the-future-from-drug-resistant-infections-en.pdf |url-status=live }}</ref> |
|||
* Accelerate progress in countries |
|||
* Innovate to secure the future |
|||
* Collaborate for more effective action |
|||
* Invest for a sustainable response |
|||
* Strengthen accountability and global governance |
|||
==Mechanisms and organisms== |
|||
===Bacteria=== |
|||
{{Further|List of antibiotic resistant bacteria}}[[File:mecA Resistance.svg|thumb|Diagram depicting antibiotic resistance through alteration of the antibiotic's target site, modeled after MRSA's resistance to penicillin. Beta-lactam antibiotics permanently inactivate [[Penicillin-binding protein|PBP enzymes]], which are essential for bacterial life, by permanently binding to their active sites. [[Methicillin-resistant Staphylococcus aureus|MRSA]], however, expresses a PBP that does not allow the antibiotic into its active site.|alt=Diagram depicting antibiotic resistance through alteration of the antibiotic's target site]] |
|||
The five main mechanisms by which bacteria exhibit resistance to antibiotics are: |
|||
# Drug inactivation or modification: for example, enzymatic deactivation of [[Penicillin|penicillin G]] in some penicillin-resistant bacteria through the production of [[Beta-lactamases|β-lactamases]]. Drugs may also be chemically modified through the addition of [[functional group]]s by [[transferase]] enzymes; for example, [[acetylation]], [[phosphorylation]], or [[adenylation]] are common resistance mechanisms to [[aminoglycoside]]s. Acetylation is the most widely used mechanism and can affect a number of [[drug class]]es.<ref name="reygaert_2018">{{cite journal | vauthors = Reygaert WC | title = An overview of the antimicrobial resistance mechanisms of bacteria | journal = AIMS Microbiology | volume = 4 | issue = 3 | pages = 482–501 | date = 2018 | pmid = 31294229 | pmc = 6604941 | doi = 10.3934/microbiol.2018.3.482 }}</ref><ref name="baylay_2019">{{cite book | vauthors = Baylay AJ, Piddock LJ, Webber MA |title=Bacterial Resistance to Antibiotics – from Molecules to Man |chapter=Molecular Mechanisms of Antibiotic Resistance – Part I |date=14 March 2019 |pages=1–26 |publisher=Wiley |doi=10.1002/9781119593522.ch1|isbn=978-1-119-94077-7 |s2cid=202806156 }}</ref>{{rp|6–8}} |
|||
# Alteration of target- or binding site: for example, alteration of [[Penicillin binding protein|PBP]]—the binding target site of penicillins—in [[Methicillin-resistant Staphylococcus aureus|MRSA]] and other penicillin-resistant bacteria. Another protective mechanism found among bacterial species is ribosomal protection proteins. These proteins protect the bacterial cell from antibiotics that target the cell's ribosomes to inhibit protein synthesis. The mechanism involves the binding of the ribosomal protection proteins to the ribosomes of the bacterial cell, which in turn changes its conformational shape. This allows the ribosomes to continue synthesizing proteins essential to the cell while preventing antibiotics from binding to the ribosome to inhibit protein synthesis.<ref>{{cite journal | vauthors = Connell SR, Tracz DM, Nierhaus KH, Taylor DE | title = Ribosomal protection proteins and their mechanism of tetracycline resistance | journal = Antimicrobial Agents and Chemotherapy | volume = 47 | issue = 12 | pages = 3675–81 | date = December 2003 | pmid = 14638464 | pmc = 296194 | doi = 10.1128/AAC.47.12.3675-3681.2003 }}</ref> |
|||
# Alteration of metabolic pathway: for example, some [[sulfa drugs|sulfonamide]]-resistant bacteria do not require [[para-aminobenzoic acid]] (PABA), an important precursor for the synthesis of [[folic acid]] and [[nucleic acid]]s in bacteria inhibited by sulfonamides, instead, like mammalian cells, they turn to using preformed folic acid.<ref>{{cite journal | vauthors = Henry RJ | title = The Mode of Action of Sulfonamides | journal = Bacteriological Reviews | volume = 7 | issue = 4 | pages = 175–262 | date = December 1943 | pmid = 16350088 | pmc = 440870 | doi = 10.1128/MMBR.7.4.175-262.1943 }}</ref> |
|||
# Reduced drug accumulation: by decreasing drug [[Semipermeable membrane|permeability]] or increasing active [[efflux (microbiology)|efflux]] (pumping out) of the drugs across the cell surface<ref>{{cite journal | vauthors = Li XZ, Nikaido H | title = Efflux-mediated drug resistance in bacteria: an update | journal = Drugs | volume = 69 | issue = 12 | pages = 1555–623 | date = August 2009 | pmid = 19678712 | pmc = 2847397 | doi = 10.2165/11317030-000000000-00000 }}</ref> These pumps within the cellular membrane of certain bacterial species are used to pump antibiotics out of the cell before they are able to do any damage. They are often activated by a specific substrate associated with an antibiotic,<ref>{{cite journal | vauthors = Aminov RI, Mackie RI | title = Evolution and ecology of antibiotic resistance genes | journal = FEMS Microbiology Letters | volume = 271 | issue = 2 | pages = 147–61 | date = June 2007 | pmid = 17490428 | doi = 10.1111/j.1574-6968.2007.00757.x | doi-access = free }}</ref> as in [[fluoroquinolone]] resistance.<ref>{{cite journal | vauthors = Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, Tsuchiya T | title = NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli | journal = Antimicrobial Agents and Chemotherapy | volume = 42 | issue = 7 | pages = 1778–82 | date = July 1998 | pmid = 9661020 | pmc = 105682 | doi = 10.1128/AAC.42.7.1778 }}</ref> |
|||
# Ribosome splitting and recycling: for example, drug-mediated stalling of the ribosome by [[lincomycin]] and [[erythromycin]] unstalled by a heat shock protein found in ''Listeria monocytogenes'', which is a homologue of HflX from other bacteria. Liberation of the ribosome from the drug allows further translation and consequent resistance to the drug.<ref>{{cite journal | vauthors = Duval M, Dar D, Carvalho F, Rocha EP, Sorek R, Cossart P | title = HflXr, a homolog of a ribosome-splitting factor, mediates antibiotic resistance | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 115 | issue = 52 | pages = 13359–13364 | date = December 2018 | pmid = 30545912 | pmc = 6310831 | doi = 10.1073/pnas.1810555115 | bibcode = 2018PNAS..11513359D | doi-access = free }}</ref> |
|||
[[File:Antibiotic resistance mechanisms.jpg|thumb|300x300px|A number of mechanisms used by common antibiotics to deal with bacteria and ways by which bacteria become resistant to them|alt=Infographic showing mechanisms for antibiotic resistance]] |
|||
There are several different types of germs that have developed a resistance over time. |
|||
The six pathogens causing most deaths associated with resistance are ''Escherichia coli'', ''Staphylococcus aureus, Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii'', and ''Pseudomonas aeruginosa''. They were responsible for 929,000 deaths attributable to resistance and 3.57 million deaths associated with resistance in 2019.<ref name="Murray_2022">{{cite journal | vauthors = Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, Han C, Bisignano C, Rao P, Wool E, Johnson SC, Browne AJ, Chipeta MG, Fell F, Hackett S, Haines-Woodhouse G, Kashef Hamadani BH, Kumaran EA, McManigal B, Achalapong S, Agarwal R, Akech S, Albertson S, Amuasi J, Andrews J, Aravkin A, Ashley E, Babin FX, Bailey F, Baker S | collaboration = Antimicrobial Resistance Collaborators | title = Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis | language = English | journal = Lancet | volume = 399 | issue = 10325 | pages = 629–655 | date = February 2022 | pmid = 35065702 | pmc = 8841637 | doi = 10.1016/S0140-6736(21)02724-0 | s2cid = 246077406 }}</ref> |
|||
Penicillinase-producing ''Neisseria gonorrhoeae'' developed a resistance to penicillin in 1976. Another example is Azithromycin-resistant ''Neisseria gonorrhoeae'', which developed a resistance to azithromycin in 2011.<ref>{{cite web|title=About Antibiotic Resistance|url=https://www.cdc.gov/drugresistance/about.html|website=CDC|date=13 March 2020|access-date=8 September 2017|archive-date=1 October 2017|archive-url=https://web.archive.org/web/20171001044758/https://www.cdc.gov/drugresistance/about.html|url-status=live}}</ref> |
|||
In [[gram-negative bacteria]], plasmid-mediated resistance genes produce proteins that can bind to [[DNA gyrase]], protecting it from the action of quinolones. Finally, mutations at key sites in DNA gyrase or [[topoisomerase IV]] can decrease their binding affinity to quinolones, decreasing the drug's effectiveness.<ref>{{cite journal | vauthors = Robicsek A, Jacoby GA, Hooper DC | title = The worldwide emergence of plasmid-mediated quinolone resistance | journal = The Lancet. Infectious Diseases | volume = 6 | issue = 10 | pages = 629–40 | date = October 2006 | pmid = 17008172 | doi = 10.1016/S1473-3099(06)70599-0 }}</ref> |
|||
Some bacteria are naturally resistant to certain antibiotics; for example, gram-negative bacteria are resistant to most [[β-lactam antibiotic]]s due to the presence of [[Beta-lactamases|β-lactamase]]. Antibiotic resistance can also be acquired as a result of either genetic mutation or [[horizontal gene transfer]].<ref>{{cite journal|vauthors=Ochiai K, Yamanaka T, Kimura K, Sawada O, O|year=1959|title=Inheritance of drug resistance (and its transfer) between Shigella strains and Between Shigella and E.coli strains|journal=Hihon Iji Shimpor|language=ja|volume=34|page=1861}}</ref> Although mutations are rare, with spontaneous mutations in the [[pathogen]] [[genome]] occurring at a rate of about 1 in 10<sup>5</sup> to 1 in 10<sup>8</sup> per chromosomal replication,<ref>{{cite book |vauthors=Watford S, Warrington SJ |chapter=Bacterial DNA Mutations |date=2018 |chapter-url=https://www.ncbi.nlm.nih.gov/books/NBK459274/ |title=StatPearls |publisher=StatPearls Publishing |pmid=29083710 |access-date=21 January 2019 |archive-date=8 March 2021 |archive-url=https://web.archive.org/web/20210308150820/https://www.ncbi.nlm.nih.gov/books/NBK459274/ |url-status=live }}</ref> the fact that bacteria reproduce at a high rate allows for the effect to be significant. Given that lifespans and production of new generations can be on a timescale of mere hours, a new (de novo) mutation in a parent cell can quickly become an [[heredity|inherited]] mutation of widespread prevalence, resulting in the [[microevolution]] of a fully resistant colony. However, chromosomal mutations also confer a cost of fitness. For example, a ribosomal mutation may protect a bacterial cell by changing the binding site of an antibiotic but may result in slower growth rate.<ref>{{cite journal | vauthors = Levin BR, Perrot V, Walker N | title = Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria | journal = Genetics | volume = 154 | issue = 3 | pages = 985–97 | date = March 2000 | doi = 10.1093/genetics/154.3.985 | pmid = 10757748 | pmc = 1460977 | url = http://www.genetics.org/cgi/pmidlookup?view=long&pmid=10757748 | access-date = 4 March 2017 | archive-date = 18 January 2023 | archive-url = https://web.archive.org/web/20230118003230/https://academic.oup.com/genetics | url-status = live }}</ref> Moreover, some adaptive mutations can propagate not only through inheritance but also through [[horizontal gene transfer]]. The most common mechanism of horizontal gene transfer is the transferring of [[Plasmid-mediated resistance|plasmids]] carrying antibiotic resistance genes between bacteria of the same or different species via [[Bacterial conjugation|conjugation]]. However, bacteria can also acquire resistance through [[Transformation (genetics)|transformation]], as in ''Streptococcus pneumoniae'' uptaking of naked fragments of extracellular DNA that contain antibiotic resistance genes to streptomycin,<ref>{{cite journal | vauthors = Hotchkiss RD | title = Transfer of penicillin resistance in pneumococci by the desoxyribonucleate derived from resistant cultures | journal = Cold Spring Harbor Symposia on Quantitative Biology | volume = 16 | pages = 457–61 | date = 1951 | pmid = 14942755 | doi = 10.1101/SQB.1951.016.01.032 }}</ref> through [[Transduction (genetics)|transduction]], as in the bacteriophage-mediated transfer of tetracycline resistance genes between strains of ''S. pyogenes'',<ref>{{cite journal | vauthors = Ubukata K, Konno M, Fujii R | title = Transduction of drug resistance to tetracycline, chloramphenicol, macrolides, lincomycin and clindamycin with phages induced from Streptococcus pyogenes | journal = The Journal of Antibiotics | volume = 28 | issue = 9 | pages = 681–8 | date = September 1975 | pmid = 1102514 | doi = 10.7164/antibiotics.28.681 | doi-access = free }}</ref> or through [[gene transfer agent]]s, which are particles produced by the host cell that resemble bacteriophage structures and are capable of transferring DNA.<ref>{{cite journal | vauthors = von Wintersdorff CJ, Penders J, van Niekerk JM, Mills ND, Majumder S, van Alphen LB, Savelkoul PH, Wolffs PF | title = Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer | journal = Frontiers in Microbiology | volume = 7 | pages = 173 | date = 19 February 2016 | pmid = 26925045 | pmc = 4759269 | doi = 10.3389/fmicb.2016.00173 | doi-access = free }}</ref> |
|||
Antibiotic resistance can be introduced artificially into a microorganism through laboratory protocols, sometimes used as a [[selectable marker]] to examine the mechanisms of gene transfer or to identify individuals that absorbed a piece of DNA that included the resistance gene and another gene of interest.<ref>{{cite journal | vauthors = Chan CX, Beiko RG, Ragan MA | title = Lateral transfer of genes and gene fragments in Staphylococcus extends beyond mobile elements | journal = Journal of Bacteriology | volume = 193 | issue = 15 | pages = 3964–77 | date = August 2011 | pmid = 21622749 | pmc = 3147504 | doi = 10.1128/JB.01524-10 }}</ref> |
|||
Recent findings show no necessity of large populations of bacteria for the appearance of antibiotic resistance. Small populations of ''[[Escherichia coli]]'' in an antibiotic gradient can become resistant. Any heterogeneous environment with respect to nutrient and antibiotic gradients may facilitate antibiotic resistance in small bacterial populations. Researchers hypothesize that the mechanism of resistance evolution is based on four SNP mutations in the genome of ''E. coli'' produced by the gradient of antibiotic.<ref>{{cite journal | vauthors = Johansen TB, Scheffer L, Jensen VK, Bohlin J, Feruglio SL | title = Whole-genome sequencing and antimicrobial resistance in Brucella melitensis from a Norwegian perspective | journal = Scientific Reports | volume = 8 | issue = 1 | pages = 8538 | date = June 2018 | pmid = 29867163 | pmc = 5986768 | doi = 10.1038/s41598-018-26906-3 | bibcode = 2018NatSR...8.8538J }}</ref> |
|||
In one study, which has implications for space microbiology, a non-pathogenic strain ''E. coli'' MG1655 was exposed to trace levels of the broad spectrum antibiotic [[chloramphenicol]], under simulated microgravity (LSMMG, or Low Shear Modeled Microgravity) over 1000 generations. The adapted strain acquired resistance to not only chloramphenicol, but also cross-resistance to other antibiotics;<ref>{{cite journal | vauthors = Tirumalai MR, Karouia F, Tran Q, Stepanov VG, Bruce RJ, Ott M, Pierson DL, Fox GE| title = Evaluation of acquired antibiotic resistance in ''Escherichia coli'' exposed to long-term low-shear modeled microgravity and background antibiotic exposure| journal = mBio | volume =10 |issue= e02637-18| date = January 2019 | pmid = 30647159 | pmc = 6336426 | doi = 10.1128/mBio.02637-18}}</ref> this was in contrast to the observation on the same strain, which was adapted to over 1000 generations under LSMMG, but without any antibiotic exposure; the strain in this case did not acquire any such resistance.<ref>{{cite journal | vauthors = Tirumalai MR, Karouia F, Tran Q, Stepanov VG, Bruce RJ, Ott M, Pierson DL, Fox GE| title = The adaptation of ''Escherichia coli'' cells grown in simulated microgravity for an extended period is both phenotypic and genomic.| journal = npj Microgravity | volume =3 |issue= 15| date = May 2017 | page = 15| pmid = 28649637 | pmc = 5460176 | doi = 10.1038/s41526-017-0020-1}}</ref> Thus, irrespective of where they are used, the use of an antibiotic would likely result in persistent resistance to that antibiotic, as well as cross-resistance to other antimicrobials. |
|||
In recent years, the emergence and spread of [[Beta-lactamases|β-lactamases]] called [[carbapenemase]]s has become a major health crisis.<ref>{{cite journal | vauthors = Ghaith DM, Mohamed ZK, Farahat MG, Aboulkasem Shahin W, Mohamed HO | title = Colonization of intestinal microbiota with carbapenemase-producing Enterobacteriaceae in paediatric intensive care units in Cairo, Egypt | journal = Arab Journal of Gastroenterology | volume = 20 | issue = 1 | pages = 19–22 | date = March 2019 | pmid = 30733176 | doi = 10.1016/j.ajg.2019.01.002 | s2cid = 73444389 | url = https://zenodo.org/record/6349599 | access-date = 30 May 2022 | archive-date = 27 November 2022 | archive-url = https://web.archive.org/web/20221127085649/https://zenodo.org/record/6349599 | url-status = live }}</ref><ref>{{cite journal | vauthors = Diene SM, Rolain JM | title = Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species | journal = Clinical Microbiology and Infection | volume = 20 | issue = 9 | pages = 831–8 | date = September 2014 | pmid = 24766097 | doi = 10.1111/1469-0691.12655 | doi-access = free }}</ref> One such carbapenemase is [[New Delhi metallo-beta-lactamase 1]] (NDM-1),<ref name="Kumarasamy">{{cite journal | vauthors = Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N | title = Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study | journal = The Lancet. Infectious Diseases | volume = 10 | issue = 9 | pages = 597–602 | date = September 2010 | pmid = 20705517 | pmc = 2933358 | doi = 10.1016/S1473-3099(10)70143-2 }}</ref> an [[enzyme]] that makes [[bacteria]] [[Antibiotic resistance|resistant]] to a broad range of [[beta-lactam antibiotic]]s. The most common bacteria that make this enzyme are gram-negative such as ''E. coli'' and ''[[Klebsiella pneumoniae]]'', but the gene for NDM-1 can spread from one strain of bacteria to another by [[horizontal gene transfer]].<ref>{{cite journal | vauthors = Hudson CM, Bent ZW, Meagher RJ, Williams KP | title = Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain | journal = PLOS ONE | volume = 9 | issue = 6 | pages = e99209 | date = 7 June 2014 | pmid = 24905728 | pmc = 4048246 | doi = 10.1371/journal.pone.0099209 | bibcode = 2014PLoSO...999209H | doi-access = free }}</ref> |
|||
===Viruses=== |
|||
Specific [[antiviral drug]]s are used to treat some viral infections. These drugs prevent viruses from reproducing by inhibiting essential stages of the virus's replication cycle in infected cells. Antivirals are used to treat [[HIV]], [[hepatitis B]], [[hepatitis C]], [[influenza]], [[herpesviridae|herpes viruses]] including [[varicella zoster virus]], [[cytomegalovirus]] and [[Epstein–Barr virus]]. With each virus, some strains have become resistant to the administered drugs.<ref>{{cite journal | vauthors = Lou Z, Sun Y, Rao Z | title = Current progress in antiviral strategies | journal = Trends in Pharmacological Sciences | volume = 35 | issue = 2 | pages = 86–102 | date = February 2014 | pmid = 24439476 | pmc = 7112804 | doi = 10.1016/j.tips.2013.11.006 }}</ref> |
|||
Antiviral drugs typically target key components of viral reproduction; for example, [[oseltamivir]] targets influenza [[neuraminidase]], while guanosine analogs inhibit viral DNA polymerase. Resistance to antivirals is thus acquired through mutations in the genes that encode the protein targets of the drugs. |
|||
Resistance to HIV antivirals is problematic, and even multi-drug resistant strains have evolved.<ref>{{cite journal | vauthors = Pennings PS | title = HIV Drug Resistance: Problems and Perspectives | journal = Infectious Disease Reports | volume = 5 | issue = Suppl 1 | pages = e5 | date = June 2013 | pmid = 24470969 | pmc = 3892620 | doi = 10.4081/idr.2013.s1.e5 }}</ref> One source of resistance is that many current HIV drugs, including NRTIs and NNRTIs, target [[reverse transcriptase]]; however, HIV-1 reverse transcriptase is highly error prone and thus mutations conferring resistance arise rapidly.<ref>{{cite journal | vauthors = Das K, Arnold E | title = HIV-1 reverse transcriptase and antiviral drug resistance. Part 1 | journal = Current Opinion in Virology | volume = 3 | issue = 2 | pages = 111–8 | date = April 2013 | pmid = 23602471 | pmc = 4097814 | doi = 10.1016/j.coviro.2013.03.012 }}</ref> Resistant strains of the HIV virus emerge rapidly if only one antiviral drug is used.<ref>{{cite journal | vauthors = Ton Q, Frenkel L | title = HIV drug resistance in mothers and infants following use of antiretrovirals to prevent mother-to-child transmission | journal = Current HIV Research | volume = 11 | issue = 2 | pages = 126–36 | date = March 2013 | pmid = 23432488 | doi = 10.2174/1570162x11311020005 }}</ref> Using three or more drugs together, termed [[combination therapy]], has helped to control this problem, but new drugs are needed because of the continuing emergence of drug-resistant HIV strains.<ref>{{cite journal | vauthors = Ebrahim O, Mazanderani AH | title = Recent developments in hiv treatment and their dissemination in poor countries | journal = Infectious Disease Reports | volume = 5 | issue = Suppl 1 | pages = e2 | date = June 2013 | pmid = 24470966 | pmc = 3892621 | doi = 10.4081/idr.2013.s1.e2 }}</ref> |
|||
===Fungi=== |
|||
Infections by fungi are a cause of high morbidity and mortality in [[Immunodeficiency|immunocompromised]] persons, such as those with HIV/AIDS, tuberculosis or receiving [[chemotherapy]].<ref>{{cite journal | vauthors = Xie JL, Polvi EJ, Shekhar-Guturja T, Cowen LE | title = Elucidating drug resistance in human fungal pathogens | journal = Future Microbiology | volume = 9 | issue = 4 | pages = 523–42 | year = 2014 | pmid = 24810351 | doi = 10.2217/fmb.14.18 }}</ref> The fungi [[Candida (fungus)|''Candida'']], ''[[Cryptococcus neoformans]]'' and ''[[Aspergillus fumigatus]]'' cause most of these infections and antifungal resistance occurs in all of them.<ref>{{cite journal | vauthors = Srinivasan A, Lopez-Ribot JL, Ramasubramanian AK | title = Overcoming antifungal resistance | journal = Drug Discovery Today: Technologies | volume = 11 | pages = 65–71 | date = March 2014 | pmid = 24847655 | pmc = 4031462 | doi = 10.1016/j.ddtec.2014.02.005 }}</ref> Multidrug resistance in fungi is increasing because of the widespread use of antifungal drugs to treat infections in immunocompromised individuals and the use of some agricultural antifungals.<ref name="Fisher_2022" /><ref>{{cite journal | vauthors = Costa C, Dias PJ, Sá-Correia I, Teixeira MC | title = MFS multidrug transporters in pathogenic fungi: do they have real clinical impact? | journal = Frontiers in Physiology | volume = 5 | pages = 197 | date = 2014 | pmid = 24904431 | pmc = 4035561 | doi = 10.3389/fphys.2014.00197 | doi-access = free }}</ref> Antifungal resistant disease is associated with increased mortality. |
|||
Some fungi (e.g. [[Candida krusei]] and [[fluconazole]]) exhibit intrinsic resistance to certain antifungal drugs or classes, whereas some species develop antifungal resistance to external pressures. Antifungal resistance is a [[One Health]] concern, driven by multiple extrinsic factors, including extensive fungicidal use, overuse of clinical antifungals, [[environmental change]] and host factors.<ref name="Fisher_2022" /> |
|||
In the USA [[fluconazole]]-resistant Candida species and azole resistance in Aspergillus fumigatus have been highlighted as a growing threat.<ref name="CDC2013" /> |
|||
More than 20 species of ''Candida'' can cause [[candidiasis]] infection, the most common of which is ''[[Candida albicans]]''. ''Candida'' yeasts normally inhabit the skin and mucous membranes without causing infection. However, overgrowth of ''Candida'' can lead to candidiasis. Some ''Candida'' species (e.g. ''[[Candida glabrata]])'' are becoming resistant to first-line and second-line [[Antifungal|antifungal agents]] such as [[echinocandin]]s and [[Azole#Use as anti-fungal agents|azoles]].<ref name="CDC2013" /> |
|||
The emergence of ''Candida auris'' as a potential human pathogen that sometimes exhibits multi-class antifungal drug resistance is concerning and has been associated with several outbreaks globally. The WHO has released a priority fungal pathogen list, including pathogens with antifungal resistance.<ref name="WHO">{{cite book |url=https://www.who.int/publications/i/item/9789240060241 |title=WHO fungal priority pathogens list to guide research, development and public health action |date=25 October 2022 |publisher=World Health Organization |editor=World Health Organization |isbn=978-92-4-006024-1 |language=En |access-date=27 October 2022 |archive-url=https://web.archive.org/web/20221026235331/https://www.who.int/publications/i/item/9789240060241 |archive-date=26 October 2022 |url-status=live}}</ref> |
|||
The identification of antifungal resistance is undermined by limited classical diagnosis of infection, where a culture is lacking, preventing susceptibility testing.<ref name="Fisher_2022" /> National and international surveillance schemes for fungal disease and antifungal resistance are limited, hampering the understanding of the disease burden and associated resistance.<ref name="Fisher_2022" /> The application of molecular testing to identify genetic markers associating with resistance may improve the identification of antifungal resistance, but the diversity of mutations associated with resistance is increasing across the fungal species causing infection. In addition, a number of resistance mechanisms depend on up-regulation of selected genes (for instance reflux pumps) rather than defined mutations that are amenable to molecular detection. |
|||
Due to the limited number of antifungals in clinical use and the increasing global incidence of antifungal resistance, using the existing antifungals in combination might be beneficial in some cases but further research is needed. Similarly, other approaches that might help to combat the emergence of antifungal resistance could rely on the development of host-directed therapies such as [[immunotherapy]] or vaccines.<ref name="Fisher_2022" /> |
|||
===Parasites=== |
|||
The [[protozoa]]n parasites that cause the diseases [[malaria]], [[trypanosomiasis]], [[toxoplasmosis]], [[cryptosporidiosis]] and [[leishmaniasis]] are important human pathogens.<ref name="pmid25057459">{{cite journal | vauthors = Andrews KT, Fisher G, Skinner-Adams TS | title = Drug repurposing and human parasitic protozoan diseases | journal = International Journal for Parasitology: Drugs and Drug Resistance | volume = 4 | issue = 2 | pages = 95–111 | date = August 2014 | pmid = 25057459 | pmc = 4095053 | doi = 10.1016/j.ijpddr.2014.02.002 }}</ref> |
|||
Malarial parasites that are resistant to the drugs that are currently available to infections are common and this has led to increased efforts to develop new drugs.<ref>{{cite journal | vauthors = Visser BJ, van Vugt M, Grobusch MP | title = Malaria: an update on current chemotherapy | journal = Expert Opinion on Pharmacotherapy | volume = 15 | issue = 15 | pages = 2219–54 | date = October 2014 | pmid = 25110058 | doi = 10.1517/14656566.2014.944499 | s2cid = 34991324 }}</ref> Resistance to recently developed drugs such as [[artemisinin]] has also been reported. The problem of drug resistance in malaria has driven efforts to develop vaccines.<ref>{{cite journal | vauthors = Chia WN, Goh YS, Rénia L | title = Novel approaches to identify protective malaria vaccine candidates | journal = Frontiers in Microbiology | volume = 5 | pages = 586 | year = 2014 | pmid = 25452745 | pmc = 4233905 | doi = 10.3389/fmicb.2014.00586 | doi-access = free }}</ref> |
|||
[[Trypanosoma|Trypanosomes]] are parasitic protozoa that cause [[African trypanosomiasis]] and [[Chagas disease]] (American trypanosomiasis).<ref>{{cite journal | vauthors = Franco JR, Simarro PP, Diarra A, Jannin JG | title = Epidemiology of human African trypanosomiasis | journal = Clinical Epidemiology | volume = 6 | pages = 257–75 | year = 2014 | pmid = 25125985 | pmc = 4130665 | doi = 10.2147/CLEP.S39728 | doi-access = free }}</ref><ref>{{cite journal | vauthors = Herrera L | title = Trypanosoma cruzi, the Causal Agent of Chagas Disease: Boundaries between Wild and Domestic Cycles in Venezuela | journal = Frontiers in Public Health | volume = 2 | pages = 259 | date = 2014 | pmid = 25506587 | pmc = 4246568 | doi = 10.3389/fpubh.2014.00259 | doi-access = free }}</ref> There are no vaccines to prevent these infections so drugs such as [[pentamidine]] and [[suramin]], [[benznidazole]] and [[nifurtimox]] are used to treat infections. These drugs are effective but infections caused by resistant parasites have been reported.<ref name="pmid25057459" /> |
|||
[[Leishmaniasis]] is caused by protozoa and is an important public health problem worldwide, especially in sub-tropical and tropical countries. Drug resistance has "become a major concern".<ref>{{cite journal | vauthors = Mansueto P, Seidita A, Vitale G, Cascio A | title = Leishmaniasis in travelers: a literature review | journal = Travel Medicine and Infectious Disease | volume = 12 | issue = 6 Pt A | pages = 563–81 | year = 2014 | pmid = 25287721 | doi = 10.1016/j.tmaid.2014.09.007 | url = https://iris.unipa.it/bitstream/10447/101959/4/Travel%20Medicine%20and%20Infectious%20Disease%202014%2012%20563-581.pdf | hdl = 10447/101959 | hdl-access = free | access-date = 23 October 2017 | archive-date = 12 October 2022 | archive-url = https://web.archive.org/web/20221012000116/https://iris.unipa.it/bitstream/10447/101959/4/Travel%20Medicine%20and%20Infectious%20Disease%202014%2012%20563-581.pdf | url-status = live }}</ref> |
|||
===Global and genomic data=== |
|||
[[File:The global resistome based on sewage-based monitoring.webp|thumb|The global 'resistome' based on sewage-based monitoring<ref name="10.1038/s41467-022-34312-7"/>]] |
|||
[[File:Gene-sharing network between bacterial genera.webp|thumb|200px|Gene-sharing network between bacterial genera<ref name="10.1038/s41467-022-34312-7"/>]] |
|||
In 2022, genomic epidemiologists reported results from a [[global health|global]] survey of antimicrobial resistance via genomic [[wastewater-based epidemiology]], finding large regional variations, providing maps, and suggesting resistance genes are also [[Horizontal gene transfer|passed on]] between microbial species that are not closely related.<ref>{{cite news |title=Antibiotika-Resistenzen verbreiten sich offenbar anders als gedacht |url=https://www.deutschlandfunknova.de/nachrichten/bakterien-antibiotika-resistenzen-verbreiten-sich-offenbar-anders-als-gedacht |access-date=17 January 2023 |work=[[Deutschlandfunk Nova]] |language=de |archive-date=17 January 2023 |archive-url=https://web.archive.org/web/20230117125129/https://www.deutschlandfunknova.de/nachrichten/bakterien-antibiotika-resistenzen-verbreiten-sich-offenbar-anders-als-gedacht |url-status=live }}</ref><ref name="10.1038/s41467-022-34312-7">{{cite journal | vauthors = Munk P, Brinch C, Møller FD, Petersen TN, Hendriksen RS, Seyfarth AM, Kjeldgaard JS, Svendsen CA, van Bunnik B, Berglund F, Larsson DG, Koopmans M, Woolhouse M, Aarestrup FM | title = Genomic analysis of sewage from 101 countries reveals global landscape of antimicrobial resistance | journal = Nature Communications | volume = 13 | issue = 1 | pages = 7251 | date = December 2022 | pmid = 36456547 | pmc = 9715550 | doi = 10.1038/s41467-022-34312-7 | doi-access = free | bibcode = 2022NatCo..13.7251M }}</ref> The [[WHO]] provides the Global Antimicrobial Resistance and Use Surveillance System (GLASS) reports which summarize annual (e.g. 2020's) data on international AMR, also including an interactive dashboard.<ref>{{cite news |title=Superbugs on the rise: WHO report signals increase in antibiotic resistance |url=https://medicalxpress.com/news/2022-12-superbugs-antibiotic-resistance.html |access-date=18 January 2023 |work=World Health Organization via medicalxpress.com|archive-date=2 February 2023 |archive-url=https://web.archive.org/web/20230202015448/https://medicalxpress.com/news/2022-12-superbugs-antibiotic-resistance.html |url-status=live }}</ref><ref>{{cite web |title=Global antimicrobial resistance and use surveillance system (GLASS) report: 2022 |url=https://www.who.int/publications/i/item/9789240062702 |website=who.int |access-date=18 January 2023|archive-date=21 January 2023 |archive-url=https://web.archive.org/web/20230121073827/https://www.who.int/publications/i/item/9789240062702 |url-status=live }}</ref> |
|||
== Epidemiology == |
|||
=== United Kingdom === |
|||
[[Public Health England]] reported that the total number of antibiotic resistant infections in England rose by 9% from 55,812 in 2017 to 60,788 in 2018, but antibiotic consumption had fallen by 9% from 20.0 to 18.2 defined daily doses per 1,000 inhabitants per day between 2014 and 2018.<ref>{{cite news |date=31 October 2019 |title=Patients contracted 165 antibiotic resistant infections each day in 2018, says PHE |publisher=Pharmaceutical Journal |url=https://www.pharmaceutical-journal.com/20207273.article |access-date=11 December 2019 |archive-date=26 July 2020 |archive-url=https://web.archive.org/web/20200726070017/https://www.pharmaceutical-journal.com/20207273.article |url-status=dead }}</ref> |
|||
===United States=== |
|||
The [[Centers for Disease Control and Prevention]] reported that more than 2.8 million cases of antibiotic resistance have been reported. However, in 2019 overall deaths from antibiotic-resistant infections decreased by 18% and deaths in hospitals decreased by 30%.<ref>{{cite web |date=2022-07-06 |title=National Infection & Death Estimates for AR |url=https://www.cdc.gov/drugresistance/national-estimates.html |access-date=2023-08-03 |website=Centers for Disease Control and Prevention|archive-date=3 August 2023 |archive-url=https://web.archive.org/web/20230803195914/https://www.cdc.gov/drugresistance/national-estimates.html |url-status=live }}</ref> |
|||
The [[COVID-19 pandemic|COVID pandemic]] caused a reversal of much of the progress made on attenuating the effects of antibiotic resistance, resulting in more antibiotic use, more resistant infections, and less data on preventive action.<ref>{{cite web |date=2022-08-11 |title=COVID-19 & Antibiotic Resistance |url=https://www.cdc.gov/drugresistance/covid19.html |access-date=2023-08-03 |website=Centers for Disease Control and Prevention|archive-date=21 February 2022 |archive-url=https://web.archive.org/web/20220221120759/https://www.cdc.gov/drugresistance/covid19.html |url-status=live }}</ref> Hospital-onset infections and deaths both increased by 15% in 2020, and significantly higher rates of infections were reported for 4 out of 6 types of healthcare associated infections.<ref>{{cite web |date=2022 |title=2022 SPECIAL REPORT: COVID-19 U.S. IMPACT ON ANTIMICROBIAL RESISTANCE |url=https://www.cdc.gov/drugresistance/pdf/covid19-impact-report-508.pdf |access-date=3 August 2023 |website=Centers for Disease Control and Prevention |page=7 |archive-date=26 August 2023 |archive-url=https://web.archive.org/web/20230826095614/https://www.cdc.gov/drugresistance/pdf/covid19-impact-report-508.pdf |url-status=live }}</ref> |
|||
== History == |
|||
The 1950s to 1970s represented the golden age of antibiotic discovery, where countless new classes of antibiotics were discovered to treat previously incurable diseases such as tuberculosis and syphilis.<ref>{{cite journal | vauthors = Aminov RI | title = A brief history of the antibiotic era: lessons learned and challenges for the future| journal = Frontiers in Microbiology | volume = 1 | pages = 134 | date = 2010 | pmid = 21687759 | pmc = 3109405 | doi = 10.3389/fmicb.2010.00134 | doi-access = free }}</ref> However, since that time the discovery of new classes of antibiotics has been almost nonexistent, and represents a situation that is especially problematic considering the resiliency of bacteria<ref>{{cite journal | vauthors = Carvalho G, Forestier C, Mathias JD | title = Antibiotic resilience: a necessary concept to complement antibiotic resistance? | journal = Proceedings. Biological Sciences | volume = 286 | issue = 1916 | pages = 20192408 | date = December 2019 | pmid = 31795866 | pmc = 6939251 | doi = 10.1098/rspb.2019.2408 }}</ref> shown over time and the continued misuse and overuse of antibiotics in treatment.<ref name="worldcat.org">{{cite book|title=Antimicrobial resistance : global report on surveillance| publisher = World Health Organization|isbn=978-92-4-156474-8|location=Geneva, Switzerland|oclc=880847527 |year=2014}}</ref> |
|||
The phenomenon of antimicrobial resistance caused by overuse of antibiotics was predicted as early as 1945 by [[Alexander Fleming]] who said "The time may come when penicillin can be bought by anyone in the shops. Then there is the danger that the ignorant man may easily under-dose himself and by exposing his microbes to nonlethal quantities of the drug make them resistant."<ref>{{cite book | veditors = Amábile-Cuevas CF | title = Antimicrobial resistance in bacteria. | publisher = Horizon Scientific Press | date = 2007 }}</ref><ref>{{Citation|url=https://www.nobelprize.org/prizes/medicine/1945/fleming/lecture/|contribution-url=https://www.nobelprize.org/uploads/2018/06/fleming-lecture.pdf|contribution=Penicillin|title=Nobel Lecture| vauthors = Fleming A |date=11 December 1945|access-date=9 August 2020|archive-url= https://web.archive.org/web/20180331001640/https://www.nobelprize.org/nobel_prizes/medicine/laureates/1945/fleming-lecture.pdf |archive-date= 31 March 2018|url-status=live}}</ref> Without the creation of new and stronger antibiotics an era where common infections and minor injuries can kill, and where complex procedures such as surgery and chemotherapy become too risky, is a very real possibility.<ref>{{cite web|url=https://www.who.int/antimicrobial-resistance/publications/global-action-plan/en/|title=WHO {{!}} Global action plan on antimicrobial resistance|website=WHO|access-date=23 April 2018|archive-url=https://web.archive.org/web/20180418062254/http://www.who.int/antimicrobial-resistance/publications/global-action-plan/en/|archive-date=18 April 2018|url-status=dead}}</ref> Antimicrobial resistance can lead to epidemics of enormous proportions if preventive actions are not taken. In this day and age current antimicrobial resistance leads to longer hospital stays, higher medical costs, and increased mortality.<ref name="worldcat.org"/> |
|||
==Society and culture== |
|||
=== Innovation policy === |
|||
Since the mid-1980s pharmaceutical companies have invested in medications for cancer or chronic disease that have greater potential to make money and have "de-emphasized or dropped development of antibiotics".<ref name="NYT_jan_21_2016" /> On 20 January 2016 at the [[World Economic Forum]] in [[Davos]], [[Switzerland]], more than "80 pharmaceutical and diagnostic companies" from around the world called for "transformational commercial models" at a global level to spur research and development on antibiotics and on the "enhanced use of diagnostic tests that can rapidly identify the infecting organism".<ref name="NYT_jan_21_2016">{{cite news | url=https://www.nytimes.com/2016/01/21/business/to-fight-superbugs-drug-makers-call-for-incentives-to-develop-antibiotics.html | title=To Fight 'Superbugs,' Drug Makers Call for Incentives to Develop Antibiotics | newspaper=New York Times |date=20 January 2016 | access-date=24 January 2016 | vauthors = Pollack A | series=Davos 2016 Special Report | location=Davos, Switzerland | archive-url=https://web.archive.org/web/20180424142407/https://www.nytimes.com/2016/01/21/business/to-fight-superbugs-drug-makers-call-for-incentives-to-develop-antibiotics.html?smid=tw-share | archive-date=24 April 2018 | url-status=live }}</ref> A number of countries are considering or implementing delinked payment models for new antimicrobials whereby payment is based on value rather than volume of drug sales. This offers the opportunity to pay for valuable new drugs even if they are reserved for use in relatively rare drug resistant infections.<ref>{{cite journal | vauthors = Leonard C, Crabb N, Glover D, Cooper S, Bouvy J, Wobbe M, Perkins M | title = Can the UK 'Netflix' Payment Model Boost the Antibacterial Pipeline? | journal = Applied Health Economics and Health Policy | volume = 21 | issue = 3 | pages = 365–372 | date = May 2023 | pmid = 36646872 | pmc = 9842493 | doi = 10.1007/s40258-022-00786-1 }}</ref> |
|||
===Legal frameworks=== |
|||
Some global health scholars have argued that a global, legal framework is needed to prevent and control antimicrobial resistance.<ref name="A. Behdinan, S.J. Hoffman 2015">{{cite journal | vauthors = Behdinan A, Hoffman SJ, Pearcey M | title = Some Global Policies for Antibiotic Resistance Depend on Legally Binding and Enforceable Commitments | journal = The Journal of Law, Medicine & Ethics | volume = 43 | issue = 2 Suppl 3 | pages = 68–73 | year = 2015 | pmid = 26243246 | doi = 10.1111/jlme.12277 | s2cid = 7415203 }}</ref><ref>{{cite journal | vauthors = Hoffman SJ, Outterson K | title = What Will It Take to Address the Global Threat of Antibiotic Resistance? | journal = The Journal of Law, Medicine & Ethics | volume = 43 | issue = 2 | pages = 363–8 | year = 2015 | pmid = 26242959 | doi = 10.1111/jlme.12253 | s2cid = 41987305 | url = https://scholarship.law.bu.edu/cgi/viewcontent.cgi?article=1007&context=faculty_scholarship | access-date = 11 November 2019 | archive-date = 12 October 2022 | archive-url = https://web.archive.org/web/20221012000131/https://scholarship.law.bu.edu/cgi/viewcontent.cgi?article=1007&context=faculty_scholarship | url-status = live }}</ref><ref name="Hoffman">{{cite journal | vauthors = Hoffman SJ, Outterson K, Røttingen JA, Cars O, Clift C, Rizvi Z, Rotberg F, Tomson G, Zorzet A | title = An international legal framework to address antimicrobial resistance | journal = Bulletin of the World Health Organization | volume = 93 | issue = 2 | pages = 66 | date = February 2015 | pmid = 25883395 | pmc = 4339972 | doi = 10.2471/BLT.15.152710 | doi-broken-date = 9 November 2024 }}</ref><ref>{{cite journal | vauthors = Rizvi Z, Hoffman SJ | title = Effective Global Action on Antibiotic Resistance Requires Careful Consideration of Convening Forums | journal = The Journal of Law, Medicine & Ethics | volume = 43 | issue = 2 Suppl 3 | pages = 74–8 | year = 2015 | pmid = 26243247 | doi = 10.1111/jlme.12278 | s2cid = 24223063 }}</ref> For instance, binding global policies could be used to create antimicrobial use standards, regulate antibiotic marketing, and strengthen global surveillance systems.<ref name="Hoffman" /><ref name="A. Behdinan, S.J. Hoffman 2015" /> Ensuring compliance of involved parties is a challenge.<ref name="Hoffman" /> Global antimicrobial resistance policies could take lessons from the environmental sector by adopting strategies that have made international environmental agreements successful in the past such as: sanctions for non-compliance, assistance for implementation, majority vote decision-making rules, an independent scientific panel, and specific commitments.<ref>{{cite journal | vauthors=Andresen S, Hoffman SJ | year=2015 | title=Much Can Be Learned about Addressing Antibiotic Resistance from Multilateral Environmental Agreements | journal=Journal of Law, Medicine & Ethics | volume=43 | issue=2| pages=46–52 }}</ref> |
|||
==== United States ==== |
|||
{{update section|date=October 2023}} |
|||
For the [[2016 United States federal budget|United States 2016 budget]], [[President of the United States|U.S. president]] [[Barack Obama]] proposed to nearly double the amount of federal funding to "combat and prevent" antibiotic resistance to more than $1.2 billion.<ref>[https://obamawhitehouse.archives.gov/the-press-office/2015/01/27/fact-sheet-president-s-2016-budget-proposes-historic-investment-combat-a President's 2016 Budget Proposes Historic Investment to Combat Antibiotic-Resistant Bacteria to Protect Public Health] {{Webarchive|url=https://web.archive.org/web/20170122010901/https://obamawhitehouse.archives.gov/the-press-office/2015/01/27/fact-sheet-president-s-2016-budget-proposes-historic-investment-combat-a |date=22 January 2017 }} The White House, Office of the Press Secretary, 27 January 2015</ref> Many international funding agencies like USAID, DFID, [[Swedish International Development Cooperation Agency|SIDA]] and [[Bill & Melinda Gates Foundation]] have pledged money for developing strategies to counter antimicrobial resistance.{{citation needed|date=April 2023}} |
|||
On 27 March 2015, the [[White House]] released a comprehensive plan to address the increasing need for agencies to combat the rise of antibiotic-resistant bacteria. The Task Force for Combating Antibiotic-Resistant Bacteria developed ''The National Action Plan for Combating Antibiotic-Resistant Bacteria'' with the intent of providing a roadmap to guide the US in the antibiotic resistance challenge and with hopes of saving many lives. This plan outlines steps taken by the Federal government over the next five years needed in order to prevent and contain outbreaks of antibiotic-resistant infections; maintain the efficacy of antibiotics already on the market; and to help to develop future diagnostics, antibiotics, and vaccines.<ref name="whitehouse.gov">{{cite web|url=https://obamawhitehouse.archives.gov/the-press-office/2015/03/27/fact-sheet-obama-administration-releases-national-action-plan-combat-ant|title=FACT SHEET: Obama Administration Releases National Action Plan to Combat Antibiotic-Resistant Bacteria|access-date=30 October 2015|archive-url=https://web.archive.org/web/20170121155651/https://obamawhitehouse.archives.gov/the-press-office/2015/03/27/fact-sheet-obama-administration-releases-national-action-plan-combat-ant|archive-date=21 January 2017|url-status=live|via=[[NARA|National Archives]]|work=[[whitehouse.gov]]|date=27 March 2015}}</ref> |
|||
The Action Plan was developed around five goals with focuses on strengthening health care, public health veterinary medicine, agriculture, food safety and research, and manufacturing. These goals, as listed by the White House, are as follows: |
|||
* Slow the Emergence of Resistant Bacteria and Prevent the Spread of Resistant Infections |
|||
* Strengthen National One-Health Surveillance Efforts to Combat Resistance |
|||
* Advance Development and use of Rapid and Innovative Diagnostic Tests for Identification and Characterization of Resistant Bacteria |
|||
* Accelerate Basic and Applied Research and Development for New Antibiotics, Other Therapeutics, and Vaccines |
|||
* Improve International Collaboration and Capacities for Antibiotic Resistance Prevention, Surveillance, Control and Antibiotic Research and Development |
|||
The following are goals set to meet by 2020:<ref name="whitehouse.gov"/> |
|||
* Establishment of antimicrobial programs within acute care hospital settings |
|||
* Reduction of inappropriate antibiotic prescription and use by at least 50% in outpatient settings and 20% inpatient settings |
|||
* Establishment of State Antibiotic Resistance (AR) Prevention Programs in all 50 states |
|||
* Elimination of the use of medically important antibiotics for growth promotion in food-producing animals. |
|||
Current Status of AMR in the U.S. |
|||
As of 2023, antimicrobial resistance (AMR) remains a significant public health threat in the United States. According to the Centers for Disease Control and Prevention's 2023 Report on Antibiotic Resistance Threats, over 2.8 million antibiotic-resistant infections occur in the U.S. each year, leading to at least 35,000 deaths annually.<ref name="CDC2023">{{cite web | title=Antibiotic Resistance Threats in the United States, 2023 | publisher=Centers for Disease Control and Prevention | url=https://www.cdc.gov/antimicrobial-resistance/data-research/threats/update-2022.html | access-date=October 20, 2023}}</ref> Among the most concerning resistant pathogens are Carbapenem-resistant Enterobacteriaceae (CRE), Methicillin-resistant Staphylococcus aureus (MRSA), and Clostridioides difficile (C. diff), all of which continue to be responsible for severe healthcare-associated infections (HAIs). |
|||
The COVID-19 pandemic led to a significant disruption in healthcare, with an increase in the use of antibiotics during the treatment of viral infections. This rise in antibiotic prescribing, coupled with overwhelmed healthcare systems, contributed to a resurgence in AMR during the pandemic years. A 2021 CDC report identified a sharp increase in HAIs caused by resistant pathogens in COVID-19 patients, a trend that has persisted into 2023.<ref name="COVID2021">{{cite journal | title=The Impact of the COVID-19 Pandemic on Antimicrobial Resistance and Management of Bloodstream Infections | journal= Pathogens| year=2023 | volume=21 | issue=6 | pages= e22–e25 | doi= 10.3390/pathogens12060780| doi-access= free| pmid= 37375470| pmc= 10302285| vauthors = Petrakis V, Panopoulou M, Rafailidis P, Lemonakis N, Lazaridis G, Terzi I, Papazoglou D, Panagopoulos P }}</ref> Recent data suggest that although antibiotic use has decreased since the pandemic, some resistant pathogens remain prevalent in healthcare settings.<ref name="CDC2023"/> |
|||
The CDC has also expanded its Get Ahead of Sepsis campaign in 2023, focusing on raising awareness of AMR's role in sepsis and promoting the judicious use of antibiotics in both healthcare and community settings.<ref name="Sepsis2023">{{cite web | title=Get Ahead of Sepsis | publisher=Centers for Disease Control and Prevention | url=https://www.cdc.gov/sepsis/get-ahead-of-sepsis/index.html | access-date=October 20, 2023}}</ref> This initiative has reached millions through social media, healthcare facilities, and public health outreach, aiming to educate the public on the importance of preventing infections and reducing antibiotic misuse. |
|||
=== Policies === |
|||
According to [[World Health Organization]], policymakers can help tackle resistance by strengthening resistance-tracking and laboratory capacity and by regulating and promoting the appropriate use of medicines.<ref name="who.int" /> Policymakers and industry can help tackle resistance by: fostering innovation and research and development of new tools; and promoting cooperation and information sharing among all stakeholders.<ref name="who.int" /> |
|||
The U.S. government continues to prioritize AMR mitigation through policy and legislation. In 2023, the National Action Plan for Combating Antibiotic-Resistant Bacteria (CARB) 2023-2028 was released, outlining strategic objectives for reducing antibiotic-resistant infections, advancing infection prevention, and accelerating research on new antibiotics.<ref name="CARB2023">{{cite web | title=National Action Plan for Combating Antibiotic-Resistant Bacteria (CARB) 2023-2028 | publisher=The White House | url=https://www.hhs.gov/sites/default/files/carb-national-action-plan-2020-2025.pdf/ | access-date=October 20, 2023}}</ref> The plan also emphasizes the importance of improving antibiotic stewardship across healthcare, agriculture, and veterinary settings. |
|||
Furthermore, the PASTEUR Act (Pioneering Antimicrobial Subscriptions to End Upsurging Resistance) has gained momentum in Congress. If passed, the bill would create a subscription-based payment model to incentivize the development of new antimicrobial drugs, while supporting antimicrobial stewardship programs to reduce the misuse of existing antibiotics.<ref name="PASTEUR">{{cite web | title=PASTEUR Act of 2023 | date=28 October 2021 | publisher=United States Congress | url=https://www.congress.gov/bill/117th-congress/house-bill/965/ | access-date=October 20, 2023}}</ref> This legislation is considered a critical step toward addressing the economic barriers to developing new antimicrobials. |
|||
=== Policy evaluation === |
|||
Measuring the costs and benefits of strategies to combat AMR is difficult and policies may only have effects in the distant future. In other infectious diseases this problem has been addressed by using mathematical models. More research is needed to understand how AMR develops and spreads so that mathematical modelling can be used to anticipate the likely effects of different policies.<ref>{{cite journal | vauthors = Knight GM, Davies NG, Colijn C, Coll F, Donker T, Gifford DR, Glover RE, Jit M, Klemm E, Lehtinen S, Lindsay JA, Lipsitch M, Llewelyn MJ, Mateus AL, Robotham JV, Sharland M, Stekel D, Yakob L, Atkins KE | title = Mathematical modelling for antibiotic resistance control policy: do we know enough? | journal = BMC Infectious Diseases | volume = 19 | issue = 1 | pages = 1011 | date = November 2019 | pmid = 31783803 | pmc = 6884858 | doi = 10.1186/s12879-019-4630-y | doi-access = free }}</ref> |
|||
== Further research == |
|||
=== Rapid testing and diagnostics === |
|||
[[File:Antibiotic sensitivity test.jpg|right|thumb|[[Antibiotic sensitivity|Antimicrobial susceptibility testing]]: Thin paper discs containing an [[antibiotic]] have been placed on an agar plate growing bacteria. Bacteria are not able to grow around antibiotics to which they are sensitive.]] |
|||
Distinguishing infections requiring antibiotics from self-limiting ones is clinically challenging. In order to guide appropriate use of antibiotics and prevent the evolution and spread of antimicrobial resistance, diagnostic tests that provide clinicians with timely, actionable results are needed. |
|||
Acute febrile illness is a common reason for seeking medical care worldwide and a major cause of morbidity and mortality. In areas with decreasing malaria incidence, many febrile patients are inappropriately treated for malaria, and in the absence of a simple diagnostic test to identify alternative causes of fever, clinicians presume that a non-malarial febrile illness is most likely a bacterial infection, leading to inappropriate use of antibiotics. Multiple studies have shown that the use of malaria rapid diagnostic tests without reliable tools to distinguish other fever causes has resulted in increased antibiotic use.<ref>{{cite journal |vauthors=Hopkins H, Bruxvoort KJ, Cairns ME, Chandler CI, Leurent B, Ansah EK, Baiden F, Baltzell KA, Björkman A, Burchett HE, Clarke SE, DiLiberto DD, Elfving K, Goodman C, Hansen KS, Kachur SP, Lal S, Lalloo DG, Leslie T, Magnussen P, Jefferies LM, Mårtensson A, Mayan I, Mbonye AK, Msellem MI, Onwujekwe OE, Owusu-Agyei S, Reyburn H, Rowland MW, Shakely D, Vestergaard LS, Webster J, Wiseman VL, Yeung S, Schellenberg D, Staedke SG, Whitty CJ |date=March 2017 |title=Impact of introduction of rapid diagnostic tests for malaria on antibiotic prescribing: analysis of observational and randomised studies in public and private healthcare settings |journal=BMJ |volume=356 |pages=j1054 |doi=10.1136/bmj.j1054 |pmc=5370398 |pmid=28356302}}</ref> |
|||
[[Antibiotic sensitivity|Antimicrobial susceptibility testing]] (AST) can facilitate a [[precision medicine]] approach to treatment by helping clinicians to prescribe more effective and targeted antimicrobial therapy.<ref>{{cite news |date=20 November 2018 |title=Diagnostics Are Helping Counter Antimicrobial Resistance, But More Work Is Needed |work=MDDI Online |url=https://www.mddionline.com/diagnostics-are-helping-counter-antimicrobial-resistance-more-work-needed |access-date=2 December 2018 |archive-date=2 December 2018 |archive-url=https://web.archive.org/web/20181202202615/https://www.mddionline.com/diagnostics-are-helping-counter-antimicrobial-resistance-more-work-needed |url-status=dead }}</ref> At the same time with traditional phenotypic AST it can take 12 to 48 hours to obtain a result due to the time taken for organisms to grow on/in culture media.<ref name="van_Belkum_2019">{{cite journal |vauthors=van Belkum A, Bachmann TT, Lüdke G, Lisby JG, Kahlmeter G, Mohess A, Becker K, Hays JP, Woodford N, Mitsakakis K, Moran-Gilad J, Vila J, Peter H, Rex JH, Dunne WM |date=January 2019 |title=Developmental roadmap for antimicrobial susceptibility testing systems |journal=Nature Reviews. Microbiology |volume=17 |issue=1 |pages=51–62 |doi=10.1038/s41579-018-0098-9 |pmc=7138758 |pmid=30333569 |doi-access=free |hdl=2445/132505}}</ref> Rapid testing, possible from [[molecular diagnostics]] innovations, is defined as "being feasible within an 8-h working shift".<ref name="van_Belkum_2019" /> There are several commercial Food and Drug Administration-approved assays available which can detect AMR genes from a variety of specimen types. Progress has been slow due to a range of reasons including cost and regulation.<ref>{{cite journal |vauthors= |date=October 2018 |title=Progress on antibiotic resistance |journal=Nature |volume=562 |issue=7727 |pages=307 |bibcode=2018Natur.562Q.307. |doi=10.1038/d41586-018-07031-7 |pmid=30333595 |doi-access=free}}</ref> Genotypic AMR characterisation methods are, however, being increasingly used in combination with machine learning algorithms in research to help better predict phenotypic AMR from organism genotype.<ref>{{cite journal |vauthors=Kim JI, Maguire F, Tsang KK, Gouliouris T, Peacock SJ, McAllister TA, McArthur AG, Beiko RG |date=September 2022 |title=Machine Learning for Antimicrobial Resistance Prediction: Current Practice, Limitations, and Clinical Perspective |journal=Clinical Microbiology Reviews |volume=35 |issue=3 |pages=e0017921 |doi=10.1128/cmr.00179-21 |pmc=9491192 |pmid=35612324 }}</ref><ref>{{cite journal |vauthors=Banerjee R, Patel R |date=February 2023 |title=Molecular diagnostics for genotypic detection of antibiotic resistance: current landscape and future directions |journal=JAC-antimicrobial Resistance |volume=5 |issue=1 |pages=dlad018 |doi=10.1093/jacamr/dlad018 |pmc=9937039 |pmid=36816746}}</ref> |
|||
Optical techniques such as phase contrast microscopy in combination with single-cell analysis are another powerful method to monitor bacterial growth. In 2017, scientists from Uppsala University in Sweden published a method<ref>{{cite journal |vauthors=Baltekin Ö, Boucharin A, Tano E, Andersson DI, Elf J |date=August 2017 |title=Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging |journal=Proceedings of the National Academy of Sciences of the United States of America |volume=114 |issue=34 |pages=9170–9175 |bibcode=2017PNAS..114.9170B |doi=10.1073/pnas.1708558114 |pmc=5576829 |pmid=28790187 |doi-access=free}}</ref> that applies principles of [[microfluidics]] and cell tracking, to monitor bacterial response to antibiotics in less than 30 minutes overall manipulation time. This invention was awarded the 8M£ Longitude Prize on AMR in 2024. Recently, this platform has been advanced by coupling microfluidic chip with [[Optical tweezers|optical tweezing]]<ref>{{cite journal |vauthors=Luro S, Potvin-Trottier L, Okumus B, Paulsson J |date=January 2020 |title=Isolating live cells after high-throughput, long-term, time-lapse microscopy |journal=Nature Methods |volume=17 |issue=1 |pages=93–100 |doi=10.1038/s41592-019-0620-7 |pmc=7525750 |pmid=31768062}}</ref> in order to isolate bacteria with altered phenotype directly from the analytical matrix. |
|||
Rapid diagnostic methods have also been trialled as antimicrobial stewardship interventions to influence the healthcare drivers of AMR. Serum [[procalcitonin]] measurement has been shown to reduce mortality rate, antimicrobial consumption and antimicrobial-related side-effects in patients with respiratory infections, but impact on AMR has not yet been demonstrated.<ref>{{cite journal |vauthors=Schuetz P, Wirz Y, Sager R, Christ-Crain M, Stolz D, Tamm M, Bouadma L, Luyt CE, Wolff M, Chastre J, Tubach F, Kristoffersen KB, Burkhardt O, Welte T, Schroeder S, Nobre V, Wei L, Bucher HC, Bhatnagar N, Annane D, Reinhart K, Branche A, Damas P, Nijsten M, de Lange DW, Deliberato RO, Lima SS, Maravić-Stojković V, Verduri A, Cao B, Shehabi Y, Beishuizen A, Jensen JS, Corti C, Van Oers JA, Falsey AR, de Jong E, Oliveira CF, Beghe B, Briel M, Mueller B |date=October 2017 |title=Procalcitonin to initiate or discontinue antibiotics in acute respiratory tract infections |journal=The Cochrane Database of Systematic Reviews |volume=10 |issue=10 |pages=CD007498 |doi=10.1002/14651858.CD007498.pub3 |pmc=6485408 |pmid=29025194 |collaboration=Cochrane Acute Respiratory Infections Group}}</ref> Similarly, point of care serum testing of the inflammatory biomarker [[C-reactive protein]] has been shown to influence antimicrobial prescribing rates in this patient cohort, but further research is required to demonstrate an effect on rates of AMR.<ref>{{cite journal |vauthors=Smedemark SA, Aabenhus R, Llor C, Fournaise A, Olsen O, Jørgensen KJ |date=October 2022 |title=Biomarkers as point-of-care tests to guide prescription of antibiotics in people with acute respiratory infections in primary care |journal=The Cochrane Database of Systematic Reviews |volume=2022 |issue=10 |pages=CD010130 |doi=10.1002/14651858.CD010130.pub3 |pmc=9575154 |pmid=36250577 |collaboration=Cochrane Acute Respiratory Infections Group}}</ref> Clinical investigation to rule out bacterial infections are often done for patients with pediatric acute respiratory infections. Currently it is unclear if rapid viral testing affects antibiotic use in children.<ref>{{cite journal |vauthors=Doan Q, Enarson P, Kissoon N, Klassen TP, Johnson DW |date=September 2014 |title=Rapid viral diagnosis for acute febrile respiratory illness in children in the Emergency Department |journal=The Cochrane Database of Systematic Reviews |volume=2014 |issue=9 |pages=CD006452 |doi=10.1002/14651858.CD006452.pub4 |pmc=6718218 |pmid=25222468}}</ref> |
|||
===Vaccines=== |
|||
Vaccines are an essential part of the response to reduce AMR as they prevent infections, reduce the use and overuse of antimicrobials, and slow the emergence and spread of drug-resistant pathogens.<ref name=WHO10October2024/> |
|||
Microorganisms usually do not develop [[Vaccine resistance|resistance to vaccines]] because vaccines reduce the spread of the infection and target the pathogen in multiple ways in the same host and possibly in different ways between different hosts. Furthermore, if the use of vaccines increases, there is evidence that antibiotic resistant strains of pathogens will decrease; the need for antibiotics will naturally decrease as vaccines prevent infection before it occurs.<ref>{{cite journal | vauthors = Mishra RP, Oviedo-Orta E, Prachi P, Rappuoli R, Bagnoli F | title = Vaccines and antibiotic resistance | journal = Current Opinion in Microbiology | volume = 15 | issue = 5 | pages = 596–602 | date = October 2012 | pmid = 22981392 | doi = 10.1016/j.mib.2012.08.002 }}</ref> A 2024 report by WHO finds that vaccines against 24 pathogens could reduce the number of antibiotics needed by 22% or 2.5 billion defined daily doses globally every year.<ref name=WHO10October2024/> If vaccines could be rolled out against all the evaluated pathogens, they could save a third of the hospital costs associated with AMR.<ref name=WHO10October2024/> Vaccinated people have fewer infections and are protected against potential complications from secondary infections that may need antimicrobial medicines or require admission to hospital.<ref name=WHO10October2024/> However, there are well documented cases of vaccine resistance, although these are usually much less of a problem than antimicrobial resistance.<ref>{{cite journal | vauthors = Kennedy DA, Read AF | title = Why does drug resistance readily evolve but vaccine resistance does not? | journal = Proceedings. Biological Sciences | volume = 284 | issue = 1851 | pages = 20162562 | date = March 2017 | pmid = 28356449 | pmc = 5378080 | doi = 10.1098/rspb.2016.2562 }}</ref><ref>{{cite journal | vauthors = Kennedy DA, Read AF | title = Why the evolution of vaccine resistance is less of a concern than the evolution of drug resistance | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 115 | issue = 51 | pages = 12878–12886 | date = December 2018 | pmid = 30559199 | pmc = 6304978 | doi = 10.1073/pnas.1717159115 | bibcode = 2018PNAS..11512878K | doi-access = free }}</ref> |
|||
While theoretically promising, antistaphylococcal vaccines have shown limited efficacy, because of immunological variation between ''Staphylococcus'' species, and the limited duration of effectiveness of the antibodies produced. Development and testing of more effective vaccines is underway.<ref>{{cite web|url=http://www.homesteadschools.com/nursing/courses/Immunity/Chapter05.html|title=Immunity, Infectious Diseases, and Pandemics—What You Can Do|publisher=HomesteadSchools.com|access-date=12 June 2013|archive-url=https://web.archive.org/web/20131203005320/http://www.homesteadschools.com/nursing/courses/Immunity/Chapter05.html|archive-date=3 December 2013|url-status=live}}</ref> |
|||
Two registrational trials have evaluated vaccine candidates in active immunization strategies against ''S. aureus'' infection. In a phase II trial, a bivalent vaccine of capsular proteins 5 & 8 was tested in 1804 hemodialysis patients with a primary fistula or synthetic graft vascular access. After 40 weeks following vaccination a protective effect was seen against ''S. aureus'' bacteremia, but not at 54 weeks following vaccination.<ref>{{cite journal | vauthors = Shinefield H, Black S, Fattom A, Horwith G, Rasgon S, Ordonez J, Yeoh H, Law D, Robbins JB, Schneerson R, Muenz L, Fuller S, Johnson J, Fireman B, Alcorn H, Naso R | title = Use of a Staphylococcus aureus conjugate vaccine in patients receiving hemodialysis | journal = The New England Journal of Medicine | volume = 346 | issue = 7 | pages = 491–6 | date = February 2002 | pmid = 11844850 | doi = 10.1056/NEJMoa011297 | doi-access = free }}</ref> Based on these results, a second trial was conducted which failed to show efficacy.<ref name="Fowler_2014">{{cite journal | vauthors = Fowler VG, Proctor RA | title = Where does a Staphylococcus aureus vaccine stand? | journal = Clinical Microbiology and Infection | volume = 20 | issue = 5 | pages = 66–75 | date = May 2014 | pmid = 24476315 | pmc = 4067250 | doi = 10.1111/1469-0691.12570 }}</ref> |
|||
Merck tested V710, a vaccine targeting IsdB, in a blinded randomized trial in patients undergoing median sternotomy. The trial was terminated after a higher rate of multiorgan system failure–related deaths was found in the V710 recipients. Vaccine recipients who developed ''S. aureus'' infection were five times more likely to die than control recipients who developed ''S. aureus'' infection.<ref>{{cite journal | vauthors = McNeely TB, Shah NA, Fridman A, Joshi A, Hartzel JS, Keshari RS, Lupu F, DiNubile MJ | title = Mortality among recipients of the Merck V710 Staphylococcus aureus vaccine after postoperative S. aureus infections: an analysis of possible contributing host factors | journal = Human Vaccines & Immunotherapeutics | volume = 10 | issue = 12 | pages = 3513–6 | date = 2014 | pmid = 25483690 | pmc = 4514053 | doi = 10.4161/hv.34407 }}</ref> |
|||
Numerous investigators have suggested that a multiple-antigen vaccine would be more effective, but a lack of biomarkers defining human protective immunity keep these proposals in the logical, but strictly hypothetical arena.<ref name="Fowler_2014" /> |
|||
===Antibody therapy=== |
|||
Antibodies are promising against antimicrobial resistance. Monoclonal antibodies (mAbs) target bacterial virulence factors, aiding in bacterial destruction through various mechanisms. Three FDA-approved antibodies target ''B. anthracis'' and ''C. difficile'' toxins.<ref>{{cite journal | vauthors = Lu RM, Hwang YC, Liu IJ, Lee CC, Tsai HZ, Li HJ, Wu HC | title = Development of therapeutic antibodies for the treatment of diseases | journal = Journal of Biomedical Science | volume = 27 | issue = 1 | pages = 1 | date = January 2020 | pmid = 31894001 | pmc = 6939334 | doi = 10.1186/s12929-019-0592-z | doi-access = free }}</ref><ref name="Singh_2024">{{cite journal | vauthors = Singh G, Rana A | title = Decoding antimicrobial resistance: unraveling molecular mechanisms and targeted strategies | journal = Archives of Microbiology | volume = 206 | issue = 6 | pages = 280 | date = May 2024 | pmid = 38805035 | doi = 10.1007/s00203-024-03998-2 | bibcode = 2024ArMic.206..280S }}</ref> Innovative strategies include DSTA4637S, an antibody-antibiotic conjugate, and MEDI13902, a bispecific antibody targeting Pseudomonas aeruginosa components.<ref name="Singh_2024" /> |
|||
===Alternating therapy=== |
|||
Alternating therapy is a proposed method in which two or three antibiotics are taken in a rotation versus taking just one antibiotic such that bacteria resistant to one antibiotic are killed when the next antibiotic is taken. Studies have found that this method reduces the rate at which antibiotic resistant bacteria emerge in vitro relative to a single drug for the entire duration.<ref>{{cite journal | vauthors = Kim S, Lieberman TD, Kishony R | title = Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 111 | issue = 40 | pages = 14494–9 | date = October 2014 | pmid = 25246554 | pmc = 4210010 | doi = 10.1073/pnas.1409800111 | bibcode = 2014PNAS..11114494K | doi-access = free }}</ref> |
|||
Studies have found that bacteria that evolve antibiotic resistance towards one group of antibiotic may become more sensitive to others.<ref>{{cite journal | vauthors = Pál C, Papp B, Lázár V | title = Collateral sensitivity of antibiotic-resistant microbes | journal = Trends in Microbiology | volume = 23 | issue = 7 | pages = 401–7 | date = July 2015 | pmid = 25818802 | pmc = 5958998 | doi = 10.1016/j.tim.2015.02.009 }}</ref> This phenomenon can be used to select against resistant bacteria using an approach termed collateral sensitivity cycling, which has recently been found to be relevant in developing treatment strategies for chronic infections caused by ''Pseudomonas aeruginosa''.<ref>{{cite journal | vauthors = Imamovic L, Ellabaan MM, Dantas Machado AM, Citterio L, Wulff T, Molin S, Krogh Johansen H, Sommer MO | title = Drug-Driven Phenotypic Convergence Supports Rational Treatment Strategies of Chronic Infections | journal = Cell | volume = 172 | issue = 1–2 | pages = 121–134.e14 | date = January 2018 | pmid = 29307490 | pmc = 5766827 | doi = 10.1016/j.cell.2017.12.012 }}</ref> Despite its promise, large-scale clinical and experimental studies revealed limited evidence of susceptibility to antibiotic cycling across various pathogens.<ref>{{cite journal | vauthors = Beckley AM, Wright ES | title = Identification of antibiotic pairs that evade concurrent resistance via a retrospective analysis of antimicrobial susceptibility test results | language = English | journal = The Lancet. Microbe | volume = 2 | issue = 10 | pages = e545–e554 | date = October 2021 | pmid = 34632433 | pmc = 8496867 | doi = 10.1016/S2666-5247(21)00118-X }}</ref><ref>{{cite journal| vauthors = Ma Y, Chua SL |date=2021-11-15|title=No collateral antibiotic sensitivity by alternating antibiotic pairs |journal=The Lancet Microbe|volume=3|issue=1 |pages=e7|language=English|doi=10.1016/S2666-5247(21)00270-6|pmid=35544116 |s2cid=244147577|issn=2666-5247|doi-access=free}}</ref> |
|||
===Development of new drugs=== |
|||
Since the discovery of antibiotics, [[research and development]] (R&D) efforts have provided new drugs in time to treat bacteria that became resistant to older antibiotics, but in the 2000s there has been concern that development has slowed enough that seriously ill people may run out of treatment options.<ref>{{cite journal | vauthors = Liu J, Bedell TA, West JG, Sorensen EJ | title = Design and Synthesis of Molecular Scaffolds with Anti-infective Activity | journal = Tetrahedron | volume = 72 | issue = 25 | pages = 3579–3592 | date = June 2016 | pmid = 27284210 | pmc = 4894353 | doi = 10.1016/j.tet.2016.01.044 }}</ref><ref>{{cite web |url= https://www.wp.dh.gov.uk/publications/files/2013/03/CMO-Annual-Report-Volume-2-20111.pdf |title=Annual Report of the Chief Medical Officer - Infections and the rise of antimicrobial resistance|date=2011|publisher=UK NHS|archive-url=https://web.archive.org/web/20131030190650/http://media.dh.gov.uk/network/357/files/2013/03/CMO-Annual-Report-Volume-2-20111.pdf|archive-date=30 October 2013|url-status=dead}}</ref> Another concern is that practitioners may become reluctant to perform routine surgeries because of the increased risk of harmful infection.<ref name="obama">{{cite news|url=https://www.npr.org/blogs/health/2013/06/04/188380562/obama-administration-seeks-to-loosen-antibiotic-approvals|title=Obama Administration Seeks To Ease Approvals For Antibiotics|newspaper=NPR.org|date=4 June 2013|publisher=NPR|access-date=7 August 2016|archive-url=https://web.archive.org/web/20150313042023/http://www.npr.org/blogs/health/2013/06/04/188380562/obama-administration-seeks-to-loosen-antibiotic-approvals|archive-date=13 March 2015|url-status=live}}</ref> Backup treatments can have serious side-effects; for example, antibiotics like [[aminoglycoside]]s (such as [[amikacin]], [[gentamicin]], [[kanamycin]], [[streptomycin]], etc.) used for the treatment of [[multi-drug-resistant tuberculosis|drug-resistant tuberculosis]] and cystic fibrosis can cause respiratory disorders, deafness and kidney failure.<ref>{{cite journal | vauthors = Prayle A, Watson A, Fortnum H, Smyth A | title = Side effects of aminoglycosides on the kidney, ear and balance in cystic fibrosis | journal = Thorax | volume = 65 | issue = 7 | pages = 654–658 | date = July 2010 | pmid = 20627927 | pmc = 2921289 | doi = 10.1136/thx.2009.131532 }}</ref><ref>{{cite journal | vauthors = Alene KA, Wangdi K, Colquhoun S, Chani K, Islam T, Rahevar K, Morishita F, Byrne A, Clark J, Viney K | title = Tuberculosis related disability: a systematic review and meta-analysis | journal = BMC Medicine | volume = 19 | issue = 1 | pages = 203 | date = September 2021 | pmid = 34496845 | pmc = 8426113 | doi = 10.1186/s12916-021-02063-9 | doi-access = free }}</ref> |
|||
The potential crisis at hand is the result of a marked decrease in industry research and development.<ref name="bbc">{{cite news|url=https://www.bbc.co.uk/news/health-21737844|title=BBC News – Antibiotics resistance 'as big a risk as terrorism' – medical chief| vauthors = Walsh F |publisher=Bbc.co.uk|access-date=12 March 2013|archive-url=https://web.archive.org/web/20180808002730/https://www.bbc.co.uk/news/health-21737844|archive-date=8 August 2018|url-status=live|work=BBC News|date=11 March 2013}}</ref><ref name=tb>{{cite journal | vauthors = Berida T, Adekunle Y, Dada-Adegbola H, Kdimy A, Roy S, Sarker S | title = Plant antibacterials: The challenges and opportunities | journal = Heliyon | volume = 10 | issue = 10 | pages = e31145 | date = May 2024 | pmid = 38803958 | pmc = 11128932 | doi = 10.1016/j.heliyon.2024.e31145 | doi-access = free | bibcode = 2024Heliy..1031145B }}</ref> Poor financial investment in antibiotic research has exacerbated the situation.<ref name="TheRealNews-2014-05-18">{{cite web | url=http://therealnews.com/t2/index.php?option=com_content&task=view&id=31&Itemid=74&jumival=11872 | title=Why Are Antibiotics Becoming Useless All Over the World? | vauthors = Khor M | author-link=Martin Khor |date=18 May 2014 | publisher=[[The Real News]] | access-date=18 May 2014 | archive-url=https://web.archive.org/web/20140518173348/http://therealnews.com/t2/index.php?option=com_content&task=view&id=31&Itemid=74&jumival=11872 | archive-date=18 May 2014 | url-status=live }}</ref><ref name="bbc" /> The pharmaceutical industry has little incentive to invest in antibiotics because of the high risk and because the potential financial returns are less likely to cover the cost of [[drug development|development]] than for other pharmaceuticals.<ref>{{cite news |title=Antibiotic Resistance: Why Aren't Drug Companies Developing New Medicines to Stop Superbugs?| vauthors = Nordrum A | website=International Business Times|year=2015}}</ref> In 2011, [[Pfizer]], one of the last major pharmaceutical companies developing new antibiotics, shut down its primary research effort, citing poor shareholder returns relative to drugs for chronic illnesses.<ref name="medpage">{{cite web|url=http://www.medpagetoday.com/InfectiousDisease/GeneralInfectiousDisease/24708|title=Pfizer Moves May Dim Prospect for New Antibiotics| vauthors = Gever J |date=4 February 2011|publisher=MedPage Today|access-date=12 March 2013 |archive-url=https://web.archive.org/web/20131214004508/http://www.medpagetoday.com/InfectiousDisease/GeneralInfectiousDisease/24708 |archive-date=14 December 2013|url-status=live}}</ref> However, small and medium-sized pharmaceutical companies are still active in antibiotic drug research. In particular, apart from classical synthetic chemistry methodologies, researchers have developed a combinatorial synthetic biology platform on single cell level in a [[high-throughput screening]] manner to diversify novel [[Lantibiotics|lanthipeptides]].<ref>{{cite journal | vauthors = Schmitt S, Montalbán-López M, Peterhoff D, Deng J, Wagner R, Held M, Kuipers OP, Panke S | title = Analysis of modular bioengineered antimicrobial lanthipeptides at nanoliter scale | journal = Nature Chemical Biology | volume = 15 | issue = 5 | pages = 437–443 | date = May 2019 | pmid = 30936500 | doi = 10.1038/s41589-019-0250-5 | s2cid = 91188986 | url = https://pure.rug.nl/ws/files/82569070/Brl_k_et_al_2019_Journal_of_Animal_Ecology.pdf | access-date = 12 April 2023 | archive-date = 18 April 2023 | archive-url = https://web.archive.org/web/20230418031100/https://pure.rug.nl/ws/files/82569070/Brl_k_et_al_2019_Journal_of_Animal_Ecology.pdf | url-status = live }}</ref> |
|||
In the 5–10 years since 2010, there has been a significant change in the ways new antimicrobial agents are discovered and developed – principally via the formation of public-private funding initiatives. These include [[CARB-X]],<ref>{{cite web |title=Overview |url=https://carb-x.org/about/overview/ |access-date=2023-03-28 |website=Carb-X|archive-date=28 March 2023 |archive-url=https://web.archive.org/web/20230328115249/https://carb-x.org/about/overview/ |url-status=live }}</ref> which focuses on nonclinical and early phase development of novel antibiotics, vaccines, rapid diagnostics; Novel Gram Negative Antibiotic (GNA-NOW),<ref>{{cite journal |title=Novel Gram Negative Antibiotic Now |url=https://cordis.europa.eu/project/id/853979 |access-date=28 March 2023 |website=CORDIS |doi=10.3030/853979 |archive-date=28 March 2023 |archive-url=https://web.archive.org/web/20230328115251/https://cordis.europa.eu/project/id/853979 |url-status=live }}</ref> which is part of the EU's [[Innovative Medicines Initiative]]; and Replenishing and Enabling the Pipeline for Anti-infective Resistance Impact Fund (REPAIR).<ref>{{cite web |title=About |url=https://www.repair-impact-fund.com/about/ |access-date=2023-03-28 |website=REPAIR Impact Fund|archive-date=23 January 2019 |archive-url=https://web.archive.org/web/20190123223739/https://www.repair-impact-fund.com/about/ |url-status=live }}</ref> Later stage clinical development is supported by the AMR Action Fund, which in turn is supported by multiple investors with the aim of developing 2–4 new antimicrobial agents by 2030. The delivery of these trials is facilitated by national and international networks supported by the Clinical Research Network of the [[National Institute for Health and Care Research]] (NIHR), European Clinical Research Alliance in Infectious Diseases (ECRAID) and the recently formed ADVANCE-ID, which is a clinical research network based in Asia.<ref>{{cite web |title=ADVANcing Clinical Evidence in Infectious Diseases (ADVANCE-ID) |url=https://sph.nus.edu.sg/2022/11/advancing-clinical-evidence-in-infectious-diseases-advance-id/ |access-date=2023-03-28 |website=sph.nus.edu.sg |archive-date=28 March 2023 |archive-url=https://web.archive.org/web/20230328115252/https://sph.nus.edu.sg/2022/11/advancing-clinical-evidence-in-infectious-diseases-advance-id/ |url-status=live }}</ref> The [[Global Antibiotic Research and Development Partnership]] (GARDP) is generating new evidence for global AMR threats such as neonatal sepsis, treatment of serious bacterial infections and sexually transmitted infections as well as addressing global access to new and strategically important antibacterial drugs.<ref>{{cite web |date=2022-09-27 |title=About GARDP |url=https://gardp.org/about-gardp/ |access-date=2023-03-28 |website=GARDP|archive-date=28 March 2023 |archive-url=https://web.archive.org/web/20230328115300/https://gardp.org/about-gardp/ |url-status=live }}</ref> |
|||
The discovery and development of new antimicrobial agents has been facilitated by regulatory advances, which have been principally led by the [[European Medicines Agency]] (EMA) and the [[Food and Drug Administration]] (FDA). These processes are increasingly aligned although important differences remain and drug developers must prepare separate documents. New development pathways have been developed to help with the approval of new antimicrobial agents that address unmet needs such as the Limited Population Pathway for Antibacterial and Antifungal Drugs (LPAD). These new pathways are required because of difficulties in conducting large definitive [[Phases of clinical research|phase III clinical trials]] in a timely way. |
|||
Some of the economic impediments to the development of new antimicrobial agents have been addressed by innovative reimbursement schemes that delink payment of antimicrobials from volume-based sales. In the UK, a market entry reward scheme has been pioneered by the [[National Institute for Health and Care Excellence|National Institute for Clinical Excellence]] (NICE) whereby an annual subscription fee is paid for use of strategically valuable antimicrobial agents – [[cefiderocol]] and [[Ceftazidime/avibactam|ceftazidime-aviabactam]] are the first agents to be used in this manner and the scheme is potential blueprint for comparable programs in other countries. |
|||
The available classes of antifungal drugs are still limited but as of 2021 novel classes of antifungals are being developed and are undergoing various stages of clinical trials to assess performance.<ref>{{cite journal | vauthors = Hoenigl M, Sprute R, Egger M, Arastehfar A, Cornely OA, Krause R, Lass-Flörl C, Prattes J, Spec A, Thompson GR, Wiederhold N, Jenks JD | title = The Antifungal Pipeline: Fosmanogepix, Ibrexafungerp, Olorofim, Opelconazole, and Rezafungin | journal = Drugs | volume = 81 | issue = 15 | pages = 1703–1729 | date = October 2021 | pmid = 34626339 | pmc = 8501344 | doi = 10.1007/s40265-021-01611-0 }}</ref> |
|||
Scientists have started using advanced computational approaches with supercomputers for the development of new antibiotic derivatives to deal with antimicrobial resistance.<ref name=tb/><ref>{{cite news |title=Antibiotic resistance outwitted by supercomputers |url=https://medicalxpress.com/news/2021-11-antibiotic-resistance-outwitted-supercomputers.html |access-date=13 December 2021 |work=[[University of Portsmouth]]|archive-date=13 December 2021 |archive-url=https://web.archive.org/web/20211213175124/https://medicalxpress.com/news/2021-11-antibiotic-resistance-outwitted-supercomputers.html |url-status=live }}</ref><ref>{{cite journal | vauthors = König G, Sokkar P, Pryk N, Heinrich S, Möller D, Cimicata G, Matzov D, Dietze P, Thiel W, Bashan A, Bandow JE, Zuegg J, Yonath A, Schulz F, Sanchez-Garcia E | title = Rational prioritization strategy allows the design of macrolide derivatives that overcome antibiotic resistance | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 118 | issue = 46 | pages = e2113632118 | date = November 2021 | pmid = 34750269 | pmc = 8609559 | doi = 10.1073/pnas.2113632118 | bibcode = 2021PNAS..11813632K | doi-access = free }}</ref> |
|||
=== Biomaterials === |
|||
Using antibiotic-free alternatives in bone infection treatment may help decrease the use of antibiotics and thus antimicrobial resistance.<ref name="The antibiotic resistance crisis: p" /> The bone regeneration material [[bioactive glass S53P4]] has shown to effectively inhibit the bacterial growth of up to 50 clinically relevant bacteria including MRSA and MRSE.<ref>{{cite journal | vauthors = Leppäranta O, Vaahtio M, Peltola T, Zhang D, Hupa L, Hupa M, Ylänen H, Salonen JI, Viljanen MK, Eerola E | title = Antibacterial effect of bioactive glasses on clinically important anaerobic bacteria in vitro | journal = Journal of Materials Science. Materials in Medicine | volume = 19 | issue = 2 | pages = 547–51 | date = February 2008 | pmid = 17619981 | doi = 10.1007/s10856-007-3018-5 | s2cid = 21444777 }}</ref><ref>{{cite journal | vauthors = Munukka E, Leppäranta O, Korkeamäki M, Vaahtio M, Peltola T, Zhang D, Hupa L, Ylänen H, Salonen JI, Viljanen MK, Eerola E | title = Bactericidal effects of bioactive glasses on clinically important aerobic bacteria | journal = Journal of Materials Science. Materials in Medicine | volume = 19 | issue = 1 | pages = 27–32 | date = January 2008 | pmid = 17569007 | doi = 10.1007/s10856-007-3143-1 | s2cid = 39643380 }}</ref><ref>{{cite journal | vauthors = Drago L, De Vecchi E, Bortolin M, Toscano M, Mattina R, Romanò CL | title = Antimicrobial activity and resistance selection of different bioglass S53P4 formulations against multidrug resistant strains | journal = Future Microbiology | volume = 10 | issue = 8 | pages = 1293–9 | date = August 2015 | pmid = 26228640 | doi = 10.2217/FMB.15.57 }}</ref> |
|||
=== Nanomaterials === |
|||
During the last decades, [[Copper nanoparticle|copper]] and [[Silver nanoparticle|silver]] [[nanomaterials]] have demonstrated appealing features for the development of a new family of antimicrobial agents.<ref>{{cite journal | vauthors = Ermini ML, Voliani V | title = Antimicrobial Nano-Agents: The Copper Age | journal = ACS Nano | volume = 15 | issue = 4 | pages = 6008–6029 | date = April 2021 | pmid = 33792292 | pmc = 8155324 | doi = 10.1021/acsnano.0c10756 | doi-access = free }}</ref> Nanoparticles (1-100 nm) show unique properties and promise as antimicrobial agents against resistant bacteria. [[Silver nanoparticle|Silver (AgNPs)]] and gold nanoparticles (AuNPs) are extensively studied, disrupting bacterial cell membranes and interfering with protein synthesis. Zinc oxide (ZnO NPs), copper (CuNPs), and silica (SiNPs) nanoparticles also exhibit antimicrobial properties. However, high synthesis costs, potential toxicity, and instability pose challenges. To overcome these, biological synthesis methods and combination therapies with other antimicrobials are explored. Enhanced biocompatibility and targeting are also under investigation to improve efficacy.<ref name="Singh_2024" /> |
|||
===Rediscovery of ancient treatments=== |
|||
Similar to the situation in malaria therapy, where successful treatments based on ancient recipes have been found,<ref>{{cite web |title=Medieval medical books could hold the recipe for new antibiotics |date=18 April 2017 |vauthors=Connelly E |work=The Conversation |url=https://theconversation.com/medieval-medical-books-could-hold-the-recipe-for-new-antibiotics-74490 |access-date=20 December 2019 |archive-date=25 December 2022 |archive-url=https://web.archive.org/web/20221225165030/http://theconversation.com/medieval-medical-books-could-hold-the-recipe-for-new-antibiotics-74490 |url-status=live }}</ref> there has already been some success in finding and testing ancient drugs and other treatments that are effective against AMR bacteria.<ref>{{cite web |url=https://www.nottingham.ac.uk/news/pressreleases/2015/march/ancientbiotics---a-medieval-remedy-for-modern-day-superbugs.aspx |title=AncientBiotics – a medieval remedy for modern day superbugs? |publisher=University of Nottingham |date=30 March 2015 |type=Press release |access-date=13 August 2019 |archive-date=21 October 2022 |archive-url=https://web.archive.org/web/20221021185808/https://www.nottingham.ac.uk/news/pressreleases/2015/march/ancientbiotics---a-medieval-remedy-for-modern-day-superbugs.aspx |url-status=live }}</ref> |
|||
=== Computational community surveillance === |
|||
One of the key tools identified by the WHO and others for the fight against rising antimicrobial resistance is improved surveillance of the spread and movement of AMR genes through different communities and regions. Recent advances in high-throughput [[DNA sequencing]] as a result of the [[Human Genome Project]] have resulted in the ability to determine the individual microbial genes in a sample.<ref name="Kim_2021">{{cite journal | vauthors = Kim DW, Cha CJ | title = Antibiotic resistome from the One-Health perspective: understanding and controlling antimicrobial resistance transmission | journal = Experimental & Molecular Medicine | volume = 53 | issue = 3 | pages = 301–309 | date = March 2021 | pmid = 33642573 | pmc = 8080597 | doi = 10.1038/s12276-021-00569-z }}</ref> Along with the availability of databases of known antimicrobial resistance genes, such as the Comprehensive Antimicrobial Resistance Database (CARD)<ref>{{cite journal | vauthors = Alcock BP, Huynh W, Chalil R, Smith KW, Raphenya AR, Wlodarski MA, Edalatmand A, Petkau A, Syed SA, Tsang KK, Baker SJ, Dave M, McCarthy MC, Mukiri KM, Nasir JA, Golbon B, Imtiaz H, Jiang X, Kaur K, Kwong M, Liang ZC, Niu KC, Shan P, Yang JY, Gray KL, Hoad GR, Jia B, Bhando T, Carfrae LA, Farha MA, French S, Gordzevich R, Rachwalski K, Tu MM, Bordeleau E, Dooley D, Griffiths E, Zubyk HL, Brown ED, Maguire F, Beiko RG, Hsiao WW, Brinkman FS, Van Domselaar G, McArthur AG | title = CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database | journal = Nucleic Acids Research | volume = 51 | issue = D1 | pages = D690–D699 | date = January 2023 | pmid = 36263822 | pmc = 9825576 | doi = 10.1093/nar/gkac920 }}</ref><ref>{{cite web |title=The Comprehensive Antibiotic Resistance Database |url=https://card.mcmaster.ca/ |access-date=2023-03-28 |website=card.mcmaster.ca |archive-date=28 March 2023 |archive-url=https://web.archive.org/web/20230328115250/https://card.mcmaster.ca/ |url-status=live }}</ref> and [[ResFinder]],<ref>{{cite journal | vauthors = Florensa AF, Kaas RS, Clausen PT, Aytan-Aktug D, Aarestrup FM | title = ResFinder – an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes | journal = Microbial Genomics | volume = 8 | issue = 1 | date = January 2022 | pmid = 35072601 | pmc = 8914360 | doi = 10.1099/mgen.0.000748 | doi-access = free }}</ref><ref>{{cite web |title=ResFinder 4.1 |url=https://cge.food.dtu.dk/services/ResFinder/ |access-date=2023-03-28 |website=cge.food.dtu.dk |archive-date=28 March 2023 |archive-url=https://web.archive.org/web/20230328115302/https://cge.food.dtu.dk/services/ResFinder/ |url-status=live }}</ref> this allows the identification of all the antimicrobial resistance genes within the sample – the so-called "[[resistome]]". In doing so, a profile of these genes within a community or environment can be determined, providing insights into how antimicrobial resistance is spreading through a population and allowing for the identification of resistance that is of concern.<ref name="Kim_2021" /> |
|||
===Phage therapy=== |
|||
{{Main|Phage therapy}} |
|||
[[Phage therapy]] is the [[therapeutic]] use of [[bacteriophage]]s to treat [[pathogenic]] [[bacterial infection]]s.<ref name="Kohn">{{cite web|url=https://www.cbsnews.com/news/silent-killers-fantastic-phages/|title=Silent Killers: Fantastic Phages?|website=[[CBS News]]|date=19 September 2002 |access-date=14 November 2017|archive-url=https://web.archive.org/web/20130210183045/http://www.cbsnews.com/stories/2002/09/19/48hours/main522596.shtml|archive-date=10 February 2013|url-status=live}}</ref> Phage therapy has many potential applications in human medicine as well as dentistry, veterinary science, and agriculture.<ref name="McAuliffe">{{cite book | chapter-url=https://www.caister.com/highveld/virology/phage.html | vauthors=McAuliffe O, Ross RP, Fitzgerald GF | chapter=The New Phage Biology: From Genomics to Applications" (introduction) | veditors=McGrath S, van Sinderen D | title=Bacteriophage: Genetics and Molecular Biology | publisher=Caister Academic Press | isbn=978-1-904455-14-1 | year=2007 | access-date=11 August 2021 | archive-date=26 November 2020 | archive-url=https://web.archive.org/web/20201126231811/https://www.caister.com/highveld/virology/phage.html | url-status=live }}</ref> |
|||
Phage therapy relies on the use of naturally occurring bacteriophages to infect and lyse bacteria at the site of infection in a host. Due to current advances in genetics and biotechnology these bacteriophages can possibly be manufactured to treat specific infections.<ref name=":2a">{{cite journal | vauthors = Lin DM, Koskella B, Lin HC | title = Phage therapy: An alternative to antibiotics in the age of multi-drug resistance | journal = World Journal of Gastrointestinal Pharmacology and Therapeutics | volume = 8 | issue = 3 | pages = 162–173 | date = August 2017 | pmid = 28828194 | pmc = 5547374 | doi = 10.4292/wjgpt.v8.i3.162 | doi-access = free }}</ref> Phages can be bioengineered to target multidrug-resistant bacterial infections, and their use involves the added benefit of preventing the elimination of beneficial bacteria in the human body.<ref name="Rather_2017" /> Phages destroy bacterial cell walls and membrane through the use of lytic proteins which kill bacteria by making many holes from the inside out.<ref name="Salmond_2015">{{cite journal | vauthors = Salmond GP, Fineran PC | title = A century of the phage: past, present and future | journal = Nature Reviews. Microbiology | volume = 13 | issue = 12 | pages = 777–86 | date = December 2015 | pmid = 26548913 | doi = 10.1038/nrmicro3564 | s2cid = 8635034 }}</ref> Bacteriophages can even possess the ability to digest the [[biofilm]] that many bacteria develop that protect them from antibiotics in order to effectively infect and kill bacteria. Bioengineering can play a role in creating successful bacteriophages.<ref name="Salmond_2015" /> |
|||
Understanding the mutual interactions and evolutions of bacterial and phage populations in the environment of a human or animal body is essential for rational phage therapy.<ref>{{cite journal | vauthors = Letarov AV, Golomidova AK, Tarasyan KK | title = Ecological basis for rational phage therapy | journal = Acta Naturae | volume = 2 | issue = 1 | pages = 60–72 | date = April 2010 | pmid = 22649629 | pmc = 3347537 | doi = 10.32607/20758251-2010-2-1-60-71 }}</ref> |
|||
[[Bacteriophage|Bacteriophagic]]s are used against antibiotic resistant bacteria in [[Georgia (country)|Georgia]] ([[George Eliava Institute]]) and in one institute in [[Wrocław]], Poland.<ref>{{cite journal | vauthors = Parfitt T | title = Georgia: an unlikely stronghold for bacteriophage therapy | journal = Lancet | volume = 365 | issue = 9478 | pages = 2166–7 | date = June 2005 | pmid = 15986542 | doi = 10.1016/S0140-6736(05)66759-1 | s2cid = 28089251 }}</ref><ref>{{cite journal | vauthors = Golkar Z, Bagasra O, Pace DG | title = Bacteriophage therapy: a potential solution for the antibiotic resistance crisis | journal = Journal of Infection in Developing Countries | volume = 8 | issue = 2 | pages = 129–36 | date = February 2014 | pmid = 24518621 | doi = 10.3855/jidc.3573 | doi-access = free }}</ref> Bacteriophage cocktails are common drugs sold over the counter in pharmacies in eastern countries.<ref name="pmid23755967">{{cite journal | vauthors = McCallin S, Alam Sarker S, Barretto C, Sultana S, Berger B, Huq S, Krause L, Bibiloni R, Schmitt B, Reuteler G, Brüssow H | title = Safety analysis of a Russian phage cocktail: from metagenomic analysis to oral application in healthy human subjects | journal = Virology | volume = 443 | issue = 2 | pages = 187–96 | date = September 2013 | pmid = 23755967 | doi = 10.1016/j.virol.2013.05.022 | doi-access = free }}</ref><ref>{{cite journal | vauthors = Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM | title = Phage treatment of human infections | journal = Bacteriophage | volume = 1 | issue = 2 | pages = 66–85 | date = March 2011 | pmid = 22334863 | pmc = 3278644 | doi = 10.4161/bact.1.2.15845 }}</ref> In Belgium, four patients with severe musculoskeletal infections received bacteriophage therapy with concomitant antibiotics. After a single course of phage therapy, no recurrence of infection occurred and no severe side-effects related to the therapy were detected.<ref>{{cite journal | vauthors = Onsea J, Soentjens P, Djebara S, Merabishvili M, Depypere M, Spriet I, De Munter P, Debaveye Y, Nijs S, Vanderschot P, Wagemans J, Pirnay JP, Lavigne R, Metsemakers WJ | title = Bacteriophage Application for Difficult-to-treat Musculoskeletal Infections: Development of a Standardized Multidisciplinary Treatment Protocol | journal = Viruses | volume = 11 | issue = 10 | page = 891 | date = September 2019 | pmid = 31548497 | pmc = 6832313 | doi = 10.3390/v11100891 | doi-access = free }}</ref> |
|||
== See also == |
|||
{{collist|colwidth=30em| |
|||
* [[Alliance for the Prudent Use of Antibiotics]] |
|||
* [[Antimicrobial resistance in Australia]] |
|||
* [[Broad-spectrum antibiotic]] |
|||
* [[Colonisation resistance]] |
|||
* [[Drug of last resort]] |
|||
* [[Genetic engineering]] |
|||
* [[Beta-lactamase#KPC (K. pneumoniae carbapenemase) (class A)|(KPC) antibacterial resistance gene]] |
|||
* [[Multidrug-resistant Gram-negative bacteria]] |
|||
* [[Multidrug-resistant tuberculosis]] |
|||
* [[New Delhi metallo-beta-lactamase 1]] |
|||
* [[Persister cells]] |
|||
* [[Resistance-nodulation-cell division superfamily]] (RND) |
|||
* [[Resistome]] |
|||
}} |
|||
== References == |
|||
<references /> |
|||
=== Books === |
|||
{{refbegin}} |
|||
* {{cite web|url=http://evolution.berkeley.edu/evolibrary/article/mutations_07|trans-title=Mutations are random|title=Understanding Evolution|publisher=University of California Museum of Paleontology|veditors=Caldwell R, Lindberg D|year=2011|access-date=15 August 2011|archive-date=8 February 2012|archive-url=https://web.archive.org/web/20120208025322/http://evolution.berkeley.edu/evolibrary/article/mutations_07|url-status=live}} |
|||
* {{cite book| veditors = Reynolds LA, Tansey EM |title=Superbugs and superdrugs : a history of MRSA : the transcript of a Witness Seminar held by the Wellcome Trust Centre for the History of Medicine at UCL, London, on 11 July 2006|date=2008|publisher=Wellcome Trust Centre for the History of Medicine at UCL|location=London|isbn=978-0-85484-114-1}} |
|||
* {{cite book | date = 2018 | doi = 10.1787/9789264307599-en | title = Stemming the Superbug Tide: Just A Few Dollars More | series = OECD Health Policy Studies | publisher = OECD Publishing | location = Paris | isbn = 978-92-64-30758-2 | s2cid = 239804815 }} |
|||
{{refend}} |
|||
=== Journals === |
|||
{{refbegin}} |
|||
* {{cite journal | vauthors = Arias CA, Murray BE | title = Antibiotic-resistant bugs in the 21st century – a clinical super-challenge | journal = The New England Journal of Medicine | volume = 360 | issue = 5 | pages = 439–443 | date = January 2009 | pmid = 19179312 | doi = 10.1056/NEJMp0804651 | s2cid = 205104375 | doi-access = free }} |
|||
* {{cite journal | title=Special Issue: Ethics and Antimicrobial Resistance | journal=Bioethics | volume=365 | issue=33 | year=2019 | url=https://onlinelibrary.wiley.com/toc/14678519/2019/33/7 | access-date=22 January 2020 | archive-date=9 March 2022 | archive-url=https://web.archive.org/web/20220309010759/https://onlinelibrary.wiley.com/toc/14678519/2019/33/7 | url-status=live }} |
|||
* {{cite journal | vauthors = Goossens H, Ferech M, Vander Stichele R, Elseviers M | title = Outpatient antibiotic use in Europe and association with resistance: a cross-national database study | journal = Lancet | volume = 365 | issue = 9459 | pages = 579–587 | year = 2005 | pmid = 15708101 | doi = 10.1016/S0140-6736(05)17907-0 | series = Group Esac Project | others = Esac Project | s2cid = 23782228 }} |
|||
* {{cite journal | vauthors = Hawkey PM, Jones AM | title = The changing epidemiology of resistance | journal = The Journal of Antimicrobial Chemotherapy | volume = 64 | issue = Suppl 1 | pages = i3–10 | date = September 2009 | pmid = 19675017 | doi = 10.1093/jac/dkp256 | url = https://academic.oup.com/jac/article-pdf/64/suppl_1/i3/2249203/dkp256.pdf | doi-access = free | access-date = 20 April 2018 | archive-date = 17 February 2024 | archive-url = https://web.archive.org/web/20240217013307/https://academic.oup.com/jac/article/64/suppl_1/i3/750430 | url-status = live }} |
|||
* {{cite journal | vauthors = Soulsby EJ | title = Resistance to antimicrobials in humans and animals | journal = BMJ | volume = 331 | issue = 7527 | pages = 1219–1220 | date = November 2005 | pmid = 16308360 | pmc = 1289307 | doi = 10.1136/bmj.331.7527.1219 }} |
|||
* {{cite journal|url=http://www.csiro.au/solutions/Alternatives-to-antibodies.html|title=Alternatives to Antibiotics Reduce Animal Disease|journal=Commonwealth Scientific and Industrial Research Organization|date=9 January 2006|access-date=26 April 2009|archive-date=5 June 2011|archive-url=https://web.archive.org/web/20110605082838/http://www.csiro.au/solutions/Alternatives-to-antibodies.html|url-status=live}} |
|||
* {{cite AV media |vauthors=Cooke P, Rees-Roberts D |year=2017 |title=CATCH |url=http://www.catchshortfilm.com/ |access-date=23 February 2017 |archive-date=9 March 2022 |archive-url=https://web.archive.org/web/20220309061945/http://www.catchshortfilm.com/ |url-status=live }} 16-minute film about a post-antibiotic world. Review: {{cite journal | vauthors = Sansom C |title=Media Watch: An intimate family story in a world without antibiotics |journal=Lancet Infect Dis |volume=17 |issue=3 |pages=274 |date=March 2017 |doi=10.1016/S1473-3099(17)30067-1 }} |
|||
{{refend}} |
|||
== Further reading == |
|||
[[File:Why won’t antibiotics cure us anymore.webm|thumb|Lecture by chemist prof. dr. Nathaniel Martin ([[Leiden University]]) on antibiotic resistance]] |
|||
* {{cite journal|author=Bancroft, EA|title=Antimicrobial resistance: it's not just for hospitals|journal=JAMA|volume=298|issue=15|pages=1803–1804|date=October 2007|doi=10.1001/jama.298.15.1803|pmid=17940239|pmc=2536104}} |
|||
* {{cite journal|author=Larson, E|title=Community factors in the development of antibiotic resistance|journal=Annual Review of Public Health|volume=28|pages=435–447|year=2007|issue=1 |pmid=17094768|doi=10.1146/annurev.publhealth.28.021406.144020|doi-access=free}} |
|||
== External links == |
|||
{{Commons category|Antimicrobial resistance}} |
|||
{{offline|med}} |
|||
* {{wikiquote-inline}} |
|||
* [https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance WHO fact sheet on antimicrobial resistance] |
|||
* [https://pharmaxchange.info/2011/02/animation-of-antimicrobial-resistance/ Animation of Antibiotic Resistance] {{Webarchive|url=https://web.archive.org/web/20220928132102/https://pharmaxchange.info/2011/02/animation-of-antimicrobial-resistance/ |date=28 September 2022 }} |
|||
* [https://www.unep.org/resources/superbugs/environmental-action Bracing for Superbugs: Strengthening environmental action in the One Health response to antimicrobial resistance] [[United Nations Environment Programme|UNEP]], 2023. |
|||
* [https://www.cdc.gov/ncidod/dhqp/pdf/ar/mdroGuideline2006.pdf CDC Guideline "Management of Multidrug-Resistant Organisms in Healthcare Settings, 2006"] |
|||
{{Antibiotics social and layman issues}} |
|||
{{Pharmacology}} |
{{Pharmacology}} |
||
{{Concepts in infectious disease}} |
|||
{{Portal bar|Biology|Medicine}} |
|||
{{DEFAULTSORT:Antibiotic Resistance}} |
{{DEFAULTSORT:Antibiotic Resistance}} |
||
[[Category: |
[[Category:Antimicrobial resistance| ]] |
||
[[Category:Evolutionary biology]] |
|||
[[Category:Health disasters]] |
[[Category:Health disasters]] |
||
[[Category:Pharmaceuticals policy]] |
[[Category:Pharmaceuticals policy]] |
||
[[Category:Veterinary medicine]] |
[[Category:Veterinary medicine]] |
||
[[Category:Global issues]] |
|||
{{Link GA|ar}} |
|||
[[ar:مقاومة المضادات الحيوية]] |
|||
[[ca:Resistència als antibiòtics]] |
|||
[[cs:Antibiotická rezistence]] |
|||
[[da:Antibiotikaresistens]] |
|||
[[de:Antibiotikum-Resistenz]] |
|||
[[es:Resistencia a antibióticos]] |
|||
[[eo:Kontraŭantibiotika rezisto]] |
|||
[[fr:Résistance aux antibiotiques]] |
|||
[[hi:प्रतिजैविक प्रतिरोध]] |
|||
[[id:Resistansi antibiotik]] |
|||
[[it:Resistenza agli antibiotici]] |
|||
[[he:עמידות לאנטיביוטיקה]] |
|||
[[hu:Antibiotikum-ellenállás]] |
|||
[[nl:Resistentie tegen antibiotica]] |
|||
[[no:Antibiotikaresistens]] |
|||
[[pl:Oporność na antybiotyki]] |
|||
[[pt:Resistência antibiótica]] |
|||
[[ro:Rezistența la antibiotice]] |
|||
[[fi:Antibioottiresistenssi]] |
|||
[[sv:Antibiotikaresistens]] |
|||
[[tr:Antibiyotik direnci]] |
|||
[[uk:Резистентність]] |
|||
Latest revision as of 22:10, 25 December 2024

Antimicrobial resistance (AMR or AR) occurs when microbes evolve mechanisms that protect them from antimicrobials, which are drugs used to treat infections.[2] This resistance affects all classes of microbes, including bacteria (antibiotic resistance), viruses (antiviral resistance), protozoa (antiprotozoal resistance), and fungi (antifungal resistance). Together, these adaptations fall under the AMR umbrella, posing significant challenges to healthcare worldwide.[3] Misuse and improper management of antimicrobials are primary drivers of this resistance, though it can also occur naturally through genetic mutations and the spread of resistant genes.[4]
Microbes resistant to multiple drugs are termed multidrug-resistant (MDR) and are sometimes called superbugs.[5] Antibiotic resistance, a significant AMR subset, enables bacteria to survive antibiotic treatment, complicating infection management and treatment options.[3] Resistance arises through spontaneous mutation, horizontal gene transfer, and increased selective pressure from antibiotic overuse, both in medicine and agriculture, which accelerates resistance development.[6]
The burden of AMR is immense, with nearly 5 million annual deaths associated with resistant infections.[7] Infections from AMR microbes are more challenging to treat and often require costly alternative therapies that may have more severe side effects.[8] Preventive measures, such as using narrow-spectrum antibiotics and improving hygiene practices, aim to reduce the spread of resistance.[9]
The WHO claims that AMR is one of the top global public health and development threats, estimating that bacterial AMR was directly responsible for 1.27 million global deaths in 2019 and contributed to 4.95 million deaths.[10] Moreover, the WHO and other international bodies warn that AMR could lead to up to 10 million deaths annually by 2050 unless actions are taken.[11] Global initiatives, such as calls for international AMR treaties, emphasize coordinated efforts to limit misuse, fund research, and provide access to necessary antimicrobials in developing nations. However, the COVID-19 pandemic redirected resources and scientific attention away from AMR, intensifying the challenge.[12]
Definition
[edit]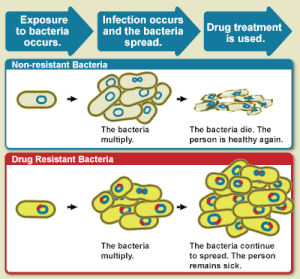
The WHO defines antimicrobial resistance as a microorganism's resistance to an antimicrobial drug that was once able to treat an infection by that microorganism.[3] A person cannot become resistant to antibiotics. Resistance is a property of the microbe, not a person or other organism infected by a microbe.[14] All types of microbes can develop drug resistance. Thus, there are antibiotic, antifungal, antiviral and antiparasitic resistance.[4][8]
Antibiotic resistance is a subset of antimicrobial resistance. This more specific resistance is linked to bacteria and thus broken down into two further subsets, microbiological and clinical. Microbiological resistance is the most common and occurs from genes, mutated or inherited, that allow the bacteria to resist the mechanism to kill the microbe associated with certain antibiotics. Clinical resistance is shown through the failure of many therapeutic techniques where the bacteria that are normally susceptible to a treatment become resistant after surviving the outcome of the treatment. In both cases of acquired resistance, the bacteria can pass the genetic catalyst for resistance through horizontal gene transfer: conjugation, transduction, or transformation. This allows the resistance to spread across the same species of pathogen or even similar bacterial pathogens.[15]
Overview
[edit]WHO report released April 2014 stated, "this serious threat is no longer a prediction for the future, it is happening right now in every region of the world and has the potential to affect anyone, of any age, in any country. Antibiotic resistance—when bacteria change so antibiotics no longer work in people who need them to treat infections—is now a major threat to public health."[16]
Each year, nearly 5 million deaths are associated with AMR globally.[7] In 2019, global deaths attributable to AMR numbered 1.27 million in 2019. That same year, AMR may have contributed to 5 million deaths and one in five people who died due to AMR were children under five years old.[17]
In 2018, WHO considered antibiotic resistance to be one of the biggest threats to global health, food security and development.[18] Deaths attributable to AMR vary by area:
| Place | Deaths per 100,000 attributable to AMR[17] |
|---|---|
| North Africa and Middle East | 11.2 |
| Southeast and East Asia, and Oceania | 11.7 |
| Latin America and Caribbean | 14.4 |
| Central and Eastern Europe and Central Asia | 17.6 |
| South Asia | 21.5 |
| Sub-Saharan Africa | 23.7 |
The European Centre for Disease Prevention and Control calculated that in 2015 there were 671,689 infections in the EU and European Economic Area caused by antibiotic-resistant bacteria, resulting in 33,110 deaths. Most were acquired in healthcare settings.[19][20] In 2019 there were 133,000 deaths caused by AMR.[21]
Causes
[edit]AMR is driven largely by the misuse and overuse of antimicrobials.[7] Yet, at the same time, many people around the world do not have access to essential antimicrobials.[7] This leads to microbes either evolving a defense against drugs used to treat them, or certain strains of microbes that have a natural resistance to antimicrobials becoming much more prevalent than the ones that are easily defeated with medication.[22] While antimicrobial resistance does occur naturally over time, the use of antimicrobial agents in a variety of settings both within the healthcare industry and outside of has led to antimicrobial resistance becoming increasingly more prevalent.[23]
Although many microbes develop resistance to antibiotics over time though natural mutation, overprescribing and inappropriate prescription of antibiotics have accelerated the problem. It is possible that as many as 1 in 3 prescriptions written for antibiotics are unnecessary.[24] Every year, approximately 154 million prescriptions for antibiotics are written. Of these, up to 46 million are unnecessary or inappropriate for the condition that the patient has.[24] Microbes may naturally develop resistance through genetic mutations that occur during cell division, and although random mutations are rare, many microbes reproduce frequently and rapidly, increasing the chances of members of the population acquiring a mutation that increases resistance.[25] Many individuals stop taking antibiotics when they begin to feel better. When this occurs, it is possible that the microbes that are less susceptible to treatment still remain in the body. If these microbes are able to continue to reproduce, this can lead to an infection by bacteria that are less susceptible or even resistant to an antibiotic.[25]
Natural occurrence
[edit]
AMR is a naturally occurring process.[2] Antimicrobial resistance can evolve naturally due to continued exposure to antimicrobials. Natural selection means that organisms that are able to adapt to their environment, survive, and continue to produce offspring.[26] As a result, the types of microorganisms that are able to survive over time with continued attack by certain antimicrobial agents will naturally become more prevalent in the environment, and those without this resistance will become obsolete.[23]
Some contemporary antimicrobial resistances have also evolved naturally before the use of antimicrobials of human clinical uses. For instance, methicillin-resistance evolved as a pathogen of hedgehogs, possibly as a co-evolutionary adaptation of the pathogen to hedgehogs that are infected by a dermatophyte that naturally produces antibiotics.[27] Also, many soil fungi and bacteria are natural competitors and the original antibiotic penicillin discovered by Alexander Fleming rapidly lost clinical effectiveness in treating humans and, furthermore, none of the other natural penicillins (F, K, N, X, O, U1 or U6) are currently in clinical use.[citation needed]
Antimicrobial resistance can be acquired from other microbes through swapping genes in a process termed horizontal gene transfer. This means that once a gene for resistance to an antibiotic appears in a microbial community, it can then spread to other microbes in the community, potentially moving from a non-disease causing microbe to a disease-causing microbe. This process is heavily driven by the natural selection processes that happen during antibiotic use or misuse.[28]
Over time, most of the strains of bacteria and infections present will be the type resistant to the antimicrobial agent being used to treat them, making this agent now ineffective to defeat most microbes. With the increased use of antimicrobial agents, there is a speeding up of this natural process.[29]
Self-medication
[edit]In the vast majority of countries, antibiotics can only be prescribed by a doctor and supplied by a pharmacy.[30] Self-medication by consumers is defined as "the taking of medicines on one's own initiative or on another person's suggestion, who is not a certified medical professional", and it has been identified as one of the primary reasons for the evolution of antimicrobial resistance.[31] Self-medication with antibiotics is an unsuitable way of using them but a common practice in resource-constrained countries. The practice exposes individuals to the risk of bacteria that have developed antimicrobial resistance.[32] Many people resort to this out of necessity, when access to a physician is unavailable, or when patients have a limited amount of time or money to see a doctor.[33] This increased access makes it extremely easy to obtain antimicrobials. An example is India, where in the state of Punjab 73% of the population resorted to treating their minor health issues and chronic illnesses through self-medication.[31]
Self-medication is higher outside the hospital environment, and this is linked to higher use of antibiotics, with the majority of antibiotics being used in the community rather than hospitals. The prevalence of self-medication in low- and middle-income countries (LMICs) ranges from 8.1% to 93%. Accessibility, affordability, and conditions of health facilities, as well as the health-seeking behavior, are factors that influence self-medication in low- and middle-income countries.[32] Two significant issues with self-medication are the lack of knowledge of the public on, firstly, the dangerous effects of certain antimicrobials (for example ciprofloxacin which can cause tendonitis, tendon rupture and aortic dissection)[34][35] and, secondly, broad microbial resistance and when to seek medical care if the infection is not clearing. In order to determine the public's knowledge and preconceived notions on antibiotic resistance, a screening of 3,537 articles published in Europe, Asia, and North America was done. Of the 55,225 total people surveyed in the articles, 70% had heard of antibiotic resistance previously, but 88% of those people thought it referred to some type of physical change in the human body.[31]
Clinical misuse
[edit]Clinical misuse by healthcare professionals is another contributor to increased antimicrobial resistance. Studies done in the US show that the indication for treatment of antibiotics, choice of the agent used, and the duration of therapy was incorrect in up to 50% of the cases studied.[36] In 2010 and 2011 about a third of antibiotic prescriptions in outpatient settings in the United States were not necessary.[37] Another study in an intensive care unit in a major hospital in France has shown that 30% to 60% of prescribed antibiotics were unnecessary.[36] These inappropriate uses of antimicrobial agents promote the evolution of antimicrobial resistance by supporting the bacteria in developing genetic alterations that lead to resistance.[38]
According to research conducted in the US that aimed to evaluate physicians' attitudes and knowledge on antimicrobial resistance in ambulatory settings, only 63% of those surveyed reported antibiotic resistance as a problem in their local practices, while 23% reported the aggressive prescription of antibiotics as necessary to avoid failing to provide adequate care.[39] This demonstrates how a majority of doctors underestimate the impact that their own prescribing habits have on antimicrobial resistance as a whole. It also confirms that some physicians may be overly cautious and prescribe antibiotics for both medical or legal reasons, even when clinical indications for use of these medications are not always confirmed. This can lead to unnecessary antimicrobial use, a pattern which may have worsened during the COVID-19 pandemic.[40][41]
Studies have shown that common misconceptions about the effectiveness and necessity of antibiotics to treat common mild illnesses contribute to their overuse.[42][43]
Important to the conversation of antibiotic use is the veterinary medical system. Veterinary oversight is required by law for all medically important antibiotics.[44] Veterinarians use the Pharmacokinetic/pharmacodynamic model (PK/PD) approach to ensuring that the correct dose of the drug is delivered to the correct place at the correct timing.[45]
Pandemics, disinfectants and healthcare systems
[edit]Increased antibiotic use during the early waves of the COVID-19 pandemic may exacerbate this global health challenge.[46][47] Moreover, pandemic burdens on some healthcare systems may contribute to antibiotic-resistant infections.[48] On the other hand, "increased hand hygiene, decreased international travel, and decreased elective hospital procedures may have reduced AMR pathogen selection and spread in the short term" during the COVID-19 pandemic.[49] The use of disinfectants such as alcohol-based hand sanitizers, and antiseptic hand wash may also have the potential to increase antimicrobial resistance.[50] Extensive use of disinfectants can lead to mutations that induce antimicrobial resistance.[51]
A 2024 United Nations High-Level Meeting on AMR has pledged to reduce deaths associated with bacterial AMR by 10% over the next six years.[7][52] In their first major declaration on the issue since 2016, global leaders also committed to raising $100 million to update and implement AMR action plans.[53] However, the final draft of the declaration omitted an earlier target to reduce antibiotic use in animals by 30% by 2030, due to opposition from meat-producing countries and the farming industry. Critics argue this omission is a major weakness, as livestock accounts for around 73% of global sales of antimicrobial agents, including antibiotics, antivirals, and antiparasitics.
Environmental pollution
[edit]Considering the complex interactions between humans, animals and the environment, it is also important to consider the environmental aspects and contributors to antimicrobial resistance.[54] Although there are still some knowledge gaps in understanding the mechanisms and transmission pathways,[55] environmental pollution is considered a significant contributor to antimicrobial resistance.[56] Important contributing factors are through "antibiotic residues", "industrial effluents", " agricultural runoffs", "heavy metals", "biocides and pesticides" and "sewage and wastewater" that create reservoirs for resistant genes and bacteria that facilitates the transfer of human pathogens.[55][56] Unused or expired antibiotics, if not disposed of properly, can enter water systems and soil.[56] Discharge from pharmaceutical manufacturing and other industrial companies can also introduce antibiotics and other chemicals into the environment.[56] These factors allow for creating selective pressure for resistant bacteria.[56] Antibiotics used in livestock and aquaculture can contaminate soil and water, which promotes resistance in environmental microbes.[55] Heavy metals such as zinc, copper and mercury, and also biocides and pesticides, can co- select for antibiotic resistance,[56] enhancing their speed.[55] Inadequate treatment of sewage and wastewater allows resistant bacteria and genes to spread through water systems.[55]
Food production
[edit]Livestock
[edit]
The antimicrobial resistance crisis also extends to the food industry, specifically with food producing animals. With an ever-increasing human population, there is constant pressure to intensify productivity in many agricultural sectors, including the production of meat as a source of protein.[57] Antibiotics are fed to livestock to act as growth supplements, and a preventive measure to decrease the likelihood of infections.[58]
Farmers typically use antibiotics in animal feed to improve growth rates and prevent infections. However, this is illogical as antibiotics are used to treat infections and not prevent infections. 80% of antibiotic use in the U.S. is for agricultural purposes and about 70% of these are medically important.[59] Overusing antibiotics gives the bacteria time to adapt leaving higher doses or even stronger antibiotics needed to combat the infection. Though antibiotics for growth promotion were banned throughout the EU in 2006, 40 countries worldwide still use antibiotics to promote growth.[60]
This can result in the transfer of resistant bacterial strains into the food that humans eat, causing potentially fatal transfer of disease. While the practice of using antibiotics as growth promoters does result in better yields and meat products, it is a major issue and needs to be decreased in order to prevent antimicrobial resistance.[61] Though the evidence linking antimicrobial usage in livestock to antimicrobial resistance is limited, the World Health Organization Advisory Group on Integrated Surveillance of Antimicrobial Resistance strongly recommended the reduction of use of medically important antimicrobials in livestock. Additionally, the Advisory Group stated that such antimicrobials should be expressly prohibited for both growth promotion and disease prevention in food producing animals.[62]
By mapping antimicrobial consumption in livestock globally, it was predicted that in 228 countries there would be a total 67% increase in consumption of antibiotics by livestock by 2030. In some countries such as Brazil, Russia, India, China, and South Africa it is predicted that a 99% increase will occur.[29] Several countries have restricted the use of antibiotics in livestock, including Canada, China, Japan, and the US. These restrictions are sometimes associated with a reduction of the prevalence of antimicrobial resistance in humans.[62]
In the United States the Veterinary Feed Directive went into practice in 2017 dictating that All medically important antibiotics to be used in feed or water for food animal species require a veterinary feed directive (VFD) or a prescription.[63]
Pesticides
[edit]Most pesticides protect crops against insects and plants, but in some cases antimicrobial pesticides are used to protect against various microorganisms such as bacteria, viruses, fungi, algae, and protozoa. The overuse of many pesticides in an effort to have a higher yield of crops has resulted in many of these microbes evolving a tolerance against these antimicrobial agents. Currently there are over 4000 antimicrobial pesticides registered with the US Environmental Protection Agency (EPA) and sold to market, showing the widespread use of these agents.[64] It is estimated that for every single meal a person consumes, 0.3 g of pesticides is used, as 90% of all pesticide use is in agriculture. A majority of these products are used to help defend against the spread of infectious diseases, and hopefully protect public health. But out of the large amount of pesticides used, it is also estimated that less than 0.1% of those antimicrobial agents, actually reach their targets. That leaves over 99% of all pesticides used available to contaminate other resources.[65] In soil, air, and water these antimicrobial agents are able to spread, coming in contact with more microorganisms and leading to these microbes evolving mechanisms to tolerate and further resist pesticides. The use of antifungal azole pesticides that drive environmental azole resistance have been linked to azole resistance cases in the clinical setting.[66] The same issues confront the novel antifungal classes (e.g. orotomides) which are again being used in both the clinic and agriculture.[67]
Wild birds
[edit]Wildlife, including wild and migratory birds, serve as a reservoir for zoonotic disease and antimicrobial-resistant organisms. Birds are a key link between the transmission of zoonotic diseases to human populations. By the same token, increased contact between wild birds and human populations (including domesticated animals), has increased the amount of anti-microbial resistance (AMR) to the bird population.[6] The introduction of AMR to wild birds positively correlates with human pollution and increased human contact. Additionally, wild birds can participate in horizontal gene transfer with bacteria, leading to the transmission of antibiotic-resistant genes (ARG).[26]
For simplicity, wild bird populations can be divided into two major categories, wild sedentary birds and wild migrating birds. Wild sedentary bird exposure to AMR is through increased contact with densely populated areas, human waste, domestic animals, and domestic animal/livestock waste. Wild migrating birds interact with sedentary birds in different environments along their migration route. This increases the rate and diversity of AMR across varying ecosystems.[6]
Neglect of wildlife in the global discussions surrounding health security and AMR, creates large barriers to true AMR surveillance. The surveillance of anti-microbial resistant organisms in wild birds is a potential metric for the rate of AMR in the environment. This surveillance also allows for further investigation into the transmission routs between different ecosystems and human populations (including domesticated animals and livestock).[6] Such information gathered from wild bird biomes, can help identify patterns of diseased transmission and better target interventions. These targeted interventions can inform the use of antimicrobial agents and reduce the persistence of multi-drug resistant organisms.[18][27]
Gene transfer from ancient microorganisms
[edit]
Permafrost is a term used to refer to any ground that remained frozen for two years or more, with the oldest known examples continuously frozen for around 700,000 years.[69] In the recent decades, permafrost has been rapidly thawing due to climate change.[70]: 1237 The cold preserves any organic matter inside the permafrost, and it is possible for microorganisms to resume their life functions once it thaws. While some common pathogens such as influenza, smallpox or the bacteria associated with pneumonia have failed to survive intentional attempts to revive them,[71] more cold-adapted microorganisms such as anthrax, or several ancient plant and amoeba viruses, have successfully survived prolonged thaw.[72][73][74][75][76]
Some scientists have argued that the inability of known causative agents of contagious diseases to survive being frozen and thawed makes this threat unlikely. Instead, there have been suggestions that when modern pathogenic bacteria interact with the ancient ones, they may, through horizontal gene transfer, pick up genetic sequences which are associated with antimicrobial resistance, exacerbating an already difficult issue.[77] Antibiotics to which permafrost bacteria have displayed at least some resistance include chloramphenicol, streptomycin, kanamycin, gentamicin, tetracycline, spectinomycin and neomycin.[78] However, other studies show that resistance levels in ancient bacteria to modern antibiotics remain lower than in the contemporary bacteria from the active layer of thawed ground above them,[68] which may mean that this risk is "no greater" than from any other soil.[79]
Prevention
[edit]
There have been increasing public calls for global collective action to address the threat, including a proposal for an international treaty on antimicrobial resistance. Further detail and attention is still needed in order to recognize and measure trends in resistance on the international level; the idea of a global tracking system has been suggested but implementation has yet to occur. A system of this nature would provide insight to areas of high resistance as well as information necessary for evaluating programs, introducing interventions and other changes made to fight or reverse antibiotic resistance.[80][81]
Duration of antimicrobials
[edit]Delaying or minimizing the use of antibiotics for certain conditions may help safely reduce their use.[82] Antimicrobial treatment duration should be based on the infection and other health problems a person may have.[83] For many infections once a person has improved there is little evidence that stopping treatment causes more resistance.[83] Some, therefore, feel that stopping early may be reasonable in some cases.[83] Other infections, however, do require long courses regardless of whether a person feels better.[83]
Delaying antibiotics for ailments such as a sore throat and otitis media may have no difference in the rate of complications compared with immediate antibiotics, for example.[82] When treating respiratory tract infections, clinical judgement is required as to the appropriate treatment (delayed or immediate antibiotic use).[82]
The study, "Shorter and Longer Antibiotic Durations for Respiratory Infections: To Fight Antimicrobial Resistance—A Retrospective Cross-Sectional Study in a Secondary Care Setting in the UK," highlights the urgency of reevaluating antibiotic treatment durations amidst the global challenge of antimicrobial resistance (AMR). It investigates the effectiveness of shorter versus longer antibiotic regimens for respiratory tract infections (RTIs) in a UK secondary care setting, emphasizing the need for evidence-based prescribing practices to optimize patient outcomes and combat AMR.[84]
Monitoring and mapping
[edit]There are multiple national and international monitoring programs for drug-resistant threats, including methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant S. aureus (VRSA), extended spectrum beta-lactamase (ESBL) producing Enterobacterales, vancomycin-resistant Enterococcus (VRE), and multidrug-resistant Acinetobacter baumannii (MRAB).[85]
ResistanceOpen is an online global map of antimicrobial resistance developed by HealthMap which displays aggregated data on antimicrobial resistance from publicly available and user submitted data.[86][87] The website can display data for a 25 miles (40 km) radius from a location. Users may submit data from antibiograms for individual hospitals or laboratories. European data is from the EARS-Net (European Antimicrobial Resistance Surveillance Network), part of the ECDC. ResistanceMap is a website by the Center for Disease Dynamics, Economics & Policy and provides data on antimicrobial resistance on a global level.[88]
The WHO's AMR global action plan also recommends antimicrobial resistance surveillance in animals.[89] Initial steps in the EU for establishing the veterinary counterpart EARS-Vet (EARS-Net for veterinary medicine) have been made.[90] AMR data from pets in particular is scarce, but needed to support antibiotic stewardship in veterinary medicine.[91]
By comparison there is a lack of national and international monitoring programs for antifungal resistance.[92]
Limiting antimicrobial use in humans
[edit]Antimicrobial stewardship programmes appear useful in reducing rates of antimicrobial resistance.[93] The antimicrobial stewardship program will also provide pharmacists with the knowledge to educate patients that antibiotics will not work for a virus for example.[94]
Excessive antimicrobial use has become one of the top contributors to the evolution of antimicrobial resistance. Since the beginning of the antimicrobial era, antimicrobials have been used to treat a wide range of infectious diseases.[95] Overuse of antimicrobials has become the primary cause of rising levels of antimicrobial resistance. The main problem is that doctors are willing to prescribe antimicrobials to ill-informed individuals who believe that antimicrobials can cure nearly all illnesses, including viral infections like the common cold. In an analysis of drug prescriptions, 36% of individuals with a cold or an upper respiratory infection (both usually viral in origin) were given prescriptions for antibiotics.[96] These prescriptions accomplished nothing other than increasing the risk of further evolution of antibiotic resistant bacteria.[97] Using antimicrobials without prescription is another driving force leading to the overuse of antibiotics to self-treat diseases like the common cold, cough, fever, and dysentery resulting in an epidemic of antibiotic resistance in countries like Bangladesh, risking its spread around the globe.[98] Introducing strict antibiotic stewardship in the outpatient setting to reduce inappropriate prescribing of antibiotics may reduce the emerging bacterial resistance.[99]
The WHO AWaRe (Access, Watch, Reserve) guidance and antibiotic book has been introduced to guide antibiotic choice for the 30 most common infections in adults and children to reduce inappropriate prescribing in primary care and hospitals. Narrow-spectrum antibiotics are preferred due to their lower resistance potential, and broad-spectrum antibiotics are only recommended for people with more severe symptoms. Some antibiotics are more likely to confer resistance, so are kept as reserve antibiotics in the AWaRe book.[100]
Various diagnostic strategies have been employed to prevent the overuse of antifungal therapy in the clinic, proving a safe alternative to empirical antifungal therapy, and thus underpinning antifungal stewardship schemes.[101]
At the hospital level
[edit]Antimicrobial stewardship teams in hospitals are encouraging optimal use of antimicrobials.[102] The goals of antimicrobial stewardship are to help practitioners pick the right drug at the right dose and duration of therapy while preventing misuse and minimizing the development of resistance. Stewardship interventions may reduce the length of stay by an average of slightly over 1 day while not increasing the risk of death.[103] Dispensing, to discharged in-house patients, the exact number of antibiotic pharmaceutical units necessary to complete an ongoing treatment can reduce antibiotic leftovers within the community as community pharmacies can have antibiotic package inefficiencies.[104]
At the primary care level
[edit]Given the volume of care provided in primary care (general practice), recent strategies have focused on reducing unnecessary antimicrobial prescribing in this setting. Simple interventions, such as written information explaining when taking antibiotics is not necessary, for example in common infections of the upper respiratory tract, have been shown to reduce antibiotic prescribing.[105] Various tools are also available to help professionals decide if prescribing antimicrobials is necessary.
Parental expectations, driven by the worry for their children's health, can influence how often children are prescribed antibiotics. Parents often rely on their clinician for advice and reassurance. However a lack of plain language information and not having adequate time for consultation negatively impacts this relationship. In effect parents often rely on past experiences in their expectations rather than reassurance from the clinician. Adequate time for consultation and plain language information can help parents make informed decisions and avoid unnecessary antibiotic use.[106]
The prescriber should closely adhere to the five rights of drug administration: the right patient, the right drug, the right dose, the right route, and the right time.[107] Microbiological samples should be taken for culture and sensitivity testing before treatment when indicated and treatment potentially changed based on the susceptibility report.[108][109]
Health workers and pharmacists can help tackle antibiotic resistance by: enhancing infection prevention and control; only prescribing and dispensing antibiotics when they are truly needed; prescribing and dispensing the right antibiotic(s) to treat the illness.[16] A unit dose system implemented in community pharmacies can also reduce antibiotic leftovers at households.[104]
At the individual level
[edit]People can help tackle resistance by using antibiotics only when infected with a bacterial infection and prescribed by a doctor; completing the full prescription even if the user is feeling better, never sharing antibiotics with others, or using leftover prescriptions.[16] Taking antibiotics when not needed won't help the user, but instead give bacteria the option to adapt and leave the user with the side effects that come with the certain type of antibiotic.[110] The CDC recommends that you follow these behaviors so that you avoid these negative side effects and keep the community safe from spreading drug-resistant bacteria.[110] Practicing basic bacterial infection prevention courses, such as hygiene, also helps to prevent the spread of antibiotic-resistant bacteria.[111]
Country examples
[edit]- The Netherlands has the lowest rate of antibiotic prescribing in the OECD, at a rate of 11.4 defined daily doses (DDD) per 1,000 people per day in 2011. The defined daily dose (DDD) is a statistical measure of drug consumption, defined by the World Health Organization (WHO).[112]
- Germany and Sweden also have lower prescribing rates, with Sweden's rate having been declining since 2007.
- Greece, France and Belgium have high prescribing rates for antibiotics of more than 28 DDD.[113]
Water, sanitation, hygiene
[edit]Infectious disease control through improved water, sanitation and hygiene (WASH) infrastructure needs to be included in the antimicrobial resistance (AMR) agenda. The "Interagency Coordination Group on Antimicrobial Resistance" stated in 2018 that "the spread of pathogens through unsafe water results in a high burden of gastrointestinal disease, increasing even further the need for antibiotic treatment."[114] This is particularly a problem in developing countries where the spread of infectious diseases caused by inadequate WASH standards is a major driver of antibiotic demand.[115] Growing usage of antibiotics together with persistent infectious disease levels have led to a dangerous cycle in which reliance on antimicrobials increases while the efficacy of drugs diminishes.[115] The proper use of infrastructure for water, sanitation and hygiene (WASH) can result in a 47–72 percent decrease of diarrhea cases treated with antibiotics depending on the type of intervention and its effectiveness.[115] A reduction of the diarrhea disease burden through improved infrastructure would result in large decreases in the number of diarrhea cases treated with antibiotics. This was estimated as ranging from 5 million in Brazil to up to 590 million in India by the year 2030.[115] The strong link between increased consumption and resistance indicates that this will directly mitigate the accelerating spread of AMR.[115] Sanitation and water for all by 2030 is Goal Number 6 of the Sustainable Development Goals.[116]
An increase in hand washing compliance by hospital staff results in decreased rates of resistant organisms.[117]
Water supply and sanitation infrastructure in health facilities offer significant co-benefits for combatting AMR, and investment should be increased.[114] There is much room for improvement: WHO and UNICEF estimated in 2015 that globally 38% of health facilities did not have a source of water, nearly 19% had no toilets and 35% had no water and soap or alcohol-based hand rub for handwashing.[118]
Industrial wastewater treatment
[edit]Manufacturers of antimicrobials need to improve the treatment of their wastewater (by using industrial wastewater treatment processes) to reduce the release of residues into the environment.[114]
Limiting antimicrobial use in animals and farming
[edit]It is established that the use of antibiotics in animal husbandry can give rise to AMR resistances in bacteria found in food animals to the antibiotics being administered (through injections or medicated feeds).[119] For this reason only antimicrobials that are deemed "not-clinically relevant" are used in these practices.
Unlike resistance to antibacterials, antifungal resistance can be driven by arable farming, currently there is no regulation on the use of similar antifungal classes in agriculture and the clinic.[92][67]
Recent studies have shown that the prophylactic use of "non-priority" or "non-clinically relevant" antimicrobials in feeds can potentially, under certain conditions, lead to co-selection of environmental AMR bacteria with resistance to medically important antibiotics.[120] The possibility for co-selection of AMR resistances in the food chain pipeline may have far-reaching implications for human health.[120][121]
Country examples
[edit]Europe
[edit]In 1997, European Union health ministers voted to ban avoparcin and four additional antibiotics used to promote animal growth in 1999.[122] In 2006 a ban on the use of antibiotics in European feed, with the exception of two antibiotics in poultry feeds, became effective.[123] In Scandinavia, there is evidence that the ban has led to a lower prevalence of antibiotic resistance in (nonhazardous) animal bacterial populations.[124] As of 2004, several European countries established a decline of antimicrobial resistance in humans through limiting the use of antimicrobials in agriculture and food industries without jeopardizing animal health or economic cost.[125]
United States
[edit]The United States Department of Agriculture (USDA) and the Food and Drug Administration (FDA) collect data on antibiotic use in humans and in a more limited fashion in animals.[126] About 80% of antibiotic use in the U.S. is for agriculture purposes, and about 70% of these are medically important.[59] This gives reason for concern about the antibiotic resistance crisis in the U.S. and more reason to monitor it. The FDA first determined in 1977 that there is evidence of emergence of antibiotic-resistant bacterial strains in livestock. The long-established practice of permitting OTC sales of antibiotics (including penicillin and other drugs) to lay animal owners for administration to their own animals nonetheless continued in all states. In 2000, the FDA announced their intention to revoke approval of fluoroquinolone use in poultry production because of substantial evidence linking it to the emergence of fluoroquinolone-resistant Campylobacter infections in humans. Legal challenges from the food animal and pharmaceutical industries delayed the final decision to do so until 2006.[127] Fluroquinolones have been banned from extra-label use in food animals in the USA since 2007.[128] However, they remain widely used in companion and exotic animals.[129]
Global action plans and awareness
[edit]At the 79th United Nations General Assembly High-Level Meeting on AMR on 26 September 2024, world leaders approved a political declaration committing to a clear set of targets and actions, including reducing the estimated 4.95 million human deaths associated with bacterial AMR annually by 10% by 2030.[7] The increasing interconnectedness of the world and the fact that new classes of antibiotics have not been developed and approved for more than 25 years highlight the extent to which antimicrobial resistance is a global health challenge.[130] A global action plan to tackle the growing problem of resistance to antibiotics and other antimicrobial medicines was endorsed at the Sixty-eighth World Health Assembly in May 2015.[89] One of the key objectives of the plan is to improve awareness and understanding of antimicrobial resistance through effective communication, education and training. This global action plan developed by the World Health Organization was created to combat the issue of antimicrobial resistance and was guided by the advice of countries and key stakeholders. The WHO's global action plan is composed of five key objectives that can be targeted through different means, and represents countries coming together to solve a major problem that can have future health consequences.[29] These objectives are as follows:
- improve awareness and understanding of antimicrobial resistance through effective communication, education and training.
- strengthen the knowledge and evidence base through surveillance and research.
- reduce the incidence of infection through effective sanitation, hygiene and infection prevention measures.
- optimize the use of antimicrobial medicines in human and animal health.
- develop the economic case for sustainable investment that takes account of the needs of all countries and to increase investment in new medicines, diagnostic tools, vaccines and other interventions.
Steps towards progress
- React based in Sweden has produced informative material on AMR for the general public.[131]
- Videos are being produced for the general public to generate interest and awareness.[132][133]
- The Irish Department of Health published a National Action Plan on Antimicrobial Resistance in October 2017.[134] The Strategy for the Control of Antimicrobial Resistance in Ireland (SARI), Iaunched in 2001 developed Guidelines for Antimicrobial Stewardship in Hospitals in Ireland[135] in conjunction with the Health Protection Surveillance Centre, these were published in 2009. Following their publication a public information campaign 'Action on Antibiotics[136]' was launched to highlight the need for a change in antibiotic prescribing. Despite this, antibiotic prescribing remains high with variance in adherence to guidelines.[137]
- The United Kingdom published a 20-year vision for antimicrobial resistance that sets out the goal of containing and controlling AMR by 2040.[138] The vision is supplemented by a 5-year action plan running from 2019 to 2024, building on the previous action plan (2013–2018).[139]
- The World Health Organization has published the 2024 Bacterial Priority Pathogens List which covers 15 families of antibiotic-resistant bacterial pathogens. Notable among these are gram-negative bacteria resistant to last-resort antibiotics, drug-resistant mycobacterium tuberculosis, and other high-burden resistant pathogens such as Salmonella, Shigella, Neisseria gonorrhoeae, Pseudomonas aeruginosa, and Staphylococcus aureus. The inclusion of these pathogens in the list underscores their global impact in terms of burden, as well as issues related to transmissibility, treatability, and prevention options. It also reflects the R&D pipeline of new treatments and emerging resistance trends.[140]
Antibiotic Awareness Week
[edit]The World Health Organization has promoted the first World Antibiotic Awareness Week running from 16 to 22 November 2015. The aim of the week is to increase global awareness of antibiotic resistance. It also wants to promote the correct usage of antibiotics across all fields in order to prevent further instances of antibiotic resistance.[141]
World Antibiotic Awareness Week has been held every November since 2015. For 2017, the Food and Agriculture Organization of the United Nations (FAO), the World Health Organization (WHO) and the World Organisation for Animal Health (OIE) are together calling for responsible use of antibiotics in humans and animals to reduce the emergence of antibiotic resistance.[142]
United Nations
In 2016 the Secretary-General of the United Nations convened the Interagency Coordination Group (IACG) on Antimicrobial Resistance.[143] The IACG worked with international organizations and experts in human, animal, and plant health to create a plan to fight antimicrobial resistance.[143] Their report released in April 2019 highlights the seriousness of antimicrobial resistance and the threat it poses to world health. It suggests five recommendations for member states to follow in order to tackle this increasing threat. The IACG recommendations are as follows:[144]
- Accelerate progress in countries
- Innovate to secure the future
- Collaborate for more effective action
- Invest for a sustainable response
- Strengthen accountability and global governance
Mechanisms and organisms
[edit]Bacteria
[edit]
The five main mechanisms by which bacteria exhibit resistance to antibiotics are:
- Drug inactivation or modification: for example, enzymatic deactivation of penicillin G in some penicillin-resistant bacteria through the production of β-lactamases. Drugs may also be chemically modified through the addition of functional groups by transferase enzymes; for example, acetylation, phosphorylation, or adenylation are common resistance mechanisms to aminoglycosides. Acetylation is the most widely used mechanism and can affect a number of drug classes.[145][146]: 6–8
- Alteration of target- or binding site: for example, alteration of PBP—the binding target site of penicillins—in MRSA and other penicillin-resistant bacteria. Another protective mechanism found among bacterial species is ribosomal protection proteins. These proteins protect the bacterial cell from antibiotics that target the cell's ribosomes to inhibit protein synthesis. The mechanism involves the binding of the ribosomal protection proteins to the ribosomes of the bacterial cell, which in turn changes its conformational shape. This allows the ribosomes to continue synthesizing proteins essential to the cell while preventing antibiotics from binding to the ribosome to inhibit protein synthesis.[147]
- Alteration of metabolic pathway: for example, some sulfonamide-resistant bacteria do not require para-aminobenzoic acid (PABA), an important precursor for the synthesis of folic acid and nucleic acids in bacteria inhibited by sulfonamides, instead, like mammalian cells, they turn to using preformed folic acid.[148]
- Reduced drug accumulation: by decreasing drug permeability or increasing active efflux (pumping out) of the drugs across the cell surface[149] These pumps within the cellular membrane of certain bacterial species are used to pump antibiotics out of the cell before they are able to do any damage. They are often activated by a specific substrate associated with an antibiotic,[150] as in fluoroquinolone resistance.[151]
- Ribosome splitting and recycling: for example, drug-mediated stalling of the ribosome by lincomycin and erythromycin unstalled by a heat shock protein found in Listeria monocytogenes, which is a homologue of HflX from other bacteria. Liberation of the ribosome from the drug allows further translation and consequent resistance to the drug.[152]
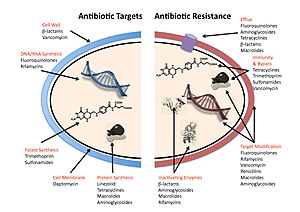
There are several different types of germs that have developed a resistance over time.
The six pathogens causing most deaths associated with resistance are Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa. They were responsible for 929,000 deaths attributable to resistance and 3.57 million deaths associated with resistance in 2019.[17]
Penicillinase-producing Neisseria gonorrhoeae developed a resistance to penicillin in 1976. Another example is Azithromycin-resistant Neisseria gonorrhoeae, which developed a resistance to azithromycin in 2011.[153]
In gram-negative bacteria, plasmid-mediated resistance genes produce proteins that can bind to DNA gyrase, protecting it from the action of quinolones. Finally, mutations at key sites in DNA gyrase or topoisomerase IV can decrease their binding affinity to quinolones, decreasing the drug's effectiveness.[154]
Some bacteria are naturally resistant to certain antibiotics; for example, gram-negative bacteria are resistant to most β-lactam antibiotics due to the presence of β-lactamase. Antibiotic resistance can also be acquired as a result of either genetic mutation or horizontal gene transfer.[155] Although mutations are rare, with spontaneous mutations in the pathogen genome occurring at a rate of about 1 in 105 to 1 in 108 per chromosomal replication,[156] the fact that bacteria reproduce at a high rate allows for the effect to be significant. Given that lifespans and production of new generations can be on a timescale of mere hours, a new (de novo) mutation in a parent cell can quickly become an inherited mutation of widespread prevalence, resulting in the microevolution of a fully resistant colony. However, chromosomal mutations also confer a cost of fitness. For example, a ribosomal mutation may protect a bacterial cell by changing the binding site of an antibiotic but may result in slower growth rate.[157] Moreover, some adaptive mutations can propagate not only through inheritance but also through horizontal gene transfer. The most common mechanism of horizontal gene transfer is the transferring of plasmids carrying antibiotic resistance genes between bacteria of the same or different species via conjugation. However, bacteria can also acquire resistance through transformation, as in Streptococcus pneumoniae uptaking of naked fragments of extracellular DNA that contain antibiotic resistance genes to streptomycin,[158] through transduction, as in the bacteriophage-mediated transfer of tetracycline resistance genes between strains of S. pyogenes,[159] or through gene transfer agents, which are particles produced by the host cell that resemble bacteriophage structures and are capable of transferring DNA.[160]
Antibiotic resistance can be introduced artificially into a microorganism through laboratory protocols, sometimes used as a selectable marker to examine the mechanisms of gene transfer or to identify individuals that absorbed a piece of DNA that included the resistance gene and another gene of interest.[161]
Recent findings show no necessity of large populations of bacteria for the appearance of antibiotic resistance. Small populations of Escherichia coli in an antibiotic gradient can become resistant. Any heterogeneous environment with respect to nutrient and antibiotic gradients may facilitate antibiotic resistance in small bacterial populations. Researchers hypothesize that the mechanism of resistance evolution is based on four SNP mutations in the genome of E. coli produced by the gradient of antibiotic.[162]
In one study, which has implications for space microbiology, a non-pathogenic strain E. coli MG1655 was exposed to trace levels of the broad spectrum antibiotic chloramphenicol, under simulated microgravity (LSMMG, or Low Shear Modeled Microgravity) over 1000 generations. The adapted strain acquired resistance to not only chloramphenicol, but also cross-resistance to other antibiotics;[163] this was in contrast to the observation on the same strain, which was adapted to over 1000 generations under LSMMG, but without any antibiotic exposure; the strain in this case did not acquire any such resistance.[164] Thus, irrespective of where they are used, the use of an antibiotic would likely result in persistent resistance to that antibiotic, as well as cross-resistance to other antimicrobials.
In recent years, the emergence and spread of β-lactamases called carbapenemases has become a major health crisis.[165][166] One such carbapenemase is New Delhi metallo-beta-lactamase 1 (NDM-1),[167] an enzyme that makes bacteria resistant to a broad range of beta-lactam antibiotics. The most common bacteria that make this enzyme are gram-negative such as E. coli and Klebsiella pneumoniae, but the gene for NDM-1 can spread from one strain of bacteria to another by horizontal gene transfer.[168]
Viruses
[edit]Specific antiviral drugs are used to treat some viral infections. These drugs prevent viruses from reproducing by inhibiting essential stages of the virus's replication cycle in infected cells. Antivirals are used to treat HIV, hepatitis B, hepatitis C, influenza, herpes viruses including varicella zoster virus, cytomegalovirus and Epstein–Barr virus. With each virus, some strains have become resistant to the administered drugs.[169]
Antiviral drugs typically target key components of viral reproduction; for example, oseltamivir targets influenza neuraminidase, while guanosine analogs inhibit viral DNA polymerase. Resistance to antivirals is thus acquired through mutations in the genes that encode the protein targets of the drugs.
Resistance to HIV antivirals is problematic, and even multi-drug resistant strains have evolved.[170] One source of resistance is that many current HIV drugs, including NRTIs and NNRTIs, target reverse transcriptase; however, HIV-1 reverse transcriptase is highly error prone and thus mutations conferring resistance arise rapidly.[171] Resistant strains of the HIV virus emerge rapidly if only one antiviral drug is used.[172] Using three or more drugs together, termed combination therapy, has helped to control this problem, but new drugs are needed because of the continuing emergence of drug-resistant HIV strains.[173]
Fungi
[edit]Infections by fungi are a cause of high morbidity and mortality in immunocompromised persons, such as those with HIV/AIDS, tuberculosis or receiving chemotherapy.[174] The fungi Candida, Cryptococcus neoformans and Aspergillus fumigatus cause most of these infections and antifungal resistance occurs in all of them.[175] Multidrug resistance in fungi is increasing because of the widespread use of antifungal drugs to treat infections in immunocompromised individuals and the use of some agricultural antifungals.[92][176] Antifungal resistant disease is associated with increased mortality.
Some fungi (e.g. Candida krusei and fluconazole) exhibit intrinsic resistance to certain antifungal drugs or classes, whereas some species develop antifungal resistance to external pressures. Antifungal resistance is a One Health concern, driven by multiple extrinsic factors, including extensive fungicidal use, overuse of clinical antifungals, environmental change and host factors.[92]
In the USA fluconazole-resistant Candida species and azole resistance in Aspergillus fumigatus have been highlighted as a growing threat.[85]
More than 20 species of Candida can cause candidiasis infection, the most common of which is Candida albicans. Candida yeasts normally inhabit the skin and mucous membranes without causing infection. However, overgrowth of Candida can lead to candidiasis. Some Candida species (e.g. Candida glabrata) are becoming resistant to first-line and second-line antifungal agents such as echinocandins and azoles.[85]
The emergence of Candida auris as a potential human pathogen that sometimes exhibits multi-class antifungal drug resistance is concerning and has been associated with several outbreaks globally. The WHO has released a priority fungal pathogen list, including pathogens with antifungal resistance.[177]
The identification of antifungal resistance is undermined by limited classical diagnosis of infection, where a culture is lacking, preventing susceptibility testing.[92] National and international surveillance schemes for fungal disease and antifungal resistance are limited, hampering the understanding of the disease burden and associated resistance.[92] The application of molecular testing to identify genetic markers associating with resistance may improve the identification of antifungal resistance, but the diversity of mutations associated with resistance is increasing across the fungal species causing infection. In addition, a number of resistance mechanisms depend on up-regulation of selected genes (for instance reflux pumps) rather than defined mutations that are amenable to molecular detection.
Due to the limited number of antifungals in clinical use and the increasing global incidence of antifungal resistance, using the existing antifungals in combination might be beneficial in some cases but further research is needed. Similarly, other approaches that might help to combat the emergence of antifungal resistance could rely on the development of host-directed therapies such as immunotherapy or vaccines.[92]
Parasites
[edit]The protozoan parasites that cause the diseases malaria, trypanosomiasis, toxoplasmosis, cryptosporidiosis and leishmaniasis are important human pathogens.[178]
Malarial parasites that are resistant to the drugs that are currently available to infections are common and this has led to increased efforts to develop new drugs.[179] Resistance to recently developed drugs such as artemisinin has also been reported. The problem of drug resistance in malaria has driven efforts to develop vaccines.[180]
Trypanosomes are parasitic protozoa that cause African trypanosomiasis and Chagas disease (American trypanosomiasis).[181][182] There are no vaccines to prevent these infections so drugs such as pentamidine and suramin, benznidazole and nifurtimox are used to treat infections. These drugs are effective but infections caused by resistant parasites have been reported.[178]
Leishmaniasis is caused by protozoa and is an important public health problem worldwide, especially in sub-tropical and tropical countries. Drug resistance has "become a major concern".[183]
Global and genomic data
[edit]

In 2022, genomic epidemiologists reported results from a global survey of antimicrobial resistance via genomic wastewater-based epidemiology, finding large regional variations, providing maps, and suggesting resistance genes are also passed on between microbial species that are not closely related.[185][184] The WHO provides the Global Antimicrobial Resistance and Use Surveillance System (GLASS) reports which summarize annual (e.g. 2020's) data on international AMR, also including an interactive dashboard.[186][187]
Epidemiology
[edit]United Kingdom
[edit]Public Health England reported that the total number of antibiotic resistant infections in England rose by 9% from 55,812 in 2017 to 60,788 in 2018, but antibiotic consumption had fallen by 9% from 20.0 to 18.2 defined daily doses per 1,000 inhabitants per day between 2014 and 2018.[188]
United States
[edit]The Centers for Disease Control and Prevention reported that more than 2.8 million cases of antibiotic resistance have been reported. However, in 2019 overall deaths from antibiotic-resistant infections decreased by 18% and deaths in hospitals decreased by 30%.[189]
The COVID pandemic caused a reversal of much of the progress made on attenuating the effects of antibiotic resistance, resulting in more antibiotic use, more resistant infections, and less data on preventive action.[190] Hospital-onset infections and deaths both increased by 15% in 2020, and significantly higher rates of infections were reported for 4 out of 6 types of healthcare associated infections.[191]
History
[edit]The 1950s to 1970s represented the golden age of antibiotic discovery, where countless new classes of antibiotics were discovered to treat previously incurable diseases such as tuberculosis and syphilis.[192] However, since that time the discovery of new classes of antibiotics has been almost nonexistent, and represents a situation that is especially problematic considering the resiliency of bacteria[193] shown over time and the continued misuse and overuse of antibiotics in treatment.[194]
The phenomenon of antimicrobial resistance caused by overuse of antibiotics was predicted as early as 1945 by Alexander Fleming who said "The time may come when penicillin can be bought by anyone in the shops. Then there is the danger that the ignorant man may easily under-dose himself and by exposing his microbes to nonlethal quantities of the drug make them resistant."[195][196] Without the creation of new and stronger antibiotics an era where common infections and minor injuries can kill, and where complex procedures such as surgery and chemotherapy become too risky, is a very real possibility.[197] Antimicrobial resistance can lead to epidemics of enormous proportions if preventive actions are not taken. In this day and age current antimicrobial resistance leads to longer hospital stays, higher medical costs, and increased mortality.[194]
Society and culture
[edit]Innovation policy
[edit]Since the mid-1980s pharmaceutical companies have invested in medications for cancer or chronic disease that have greater potential to make money and have "de-emphasized or dropped development of antibiotics".[198] On 20 January 2016 at the World Economic Forum in Davos, Switzerland, more than "80 pharmaceutical and diagnostic companies" from around the world called for "transformational commercial models" at a global level to spur research and development on antibiotics and on the "enhanced use of diagnostic tests that can rapidly identify the infecting organism".[198] A number of countries are considering or implementing delinked payment models for new antimicrobials whereby payment is based on value rather than volume of drug sales. This offers the opportunity to pay for valuable new drugs even if they are reserved for use in relatively rare drug resistant infections.[199]
Legal frameworks
[edit]Some global health scholars have argued that a global, legal framework is needed to prevent and control antimicrobial resistance.[200][201][202][203] For instance, binding global policies could be used to create antimicrobial use standards, regulate antibiotic marketing, and strengthen global surveillance systems.[202][200] Ensuring compliance of involved parties is a challenge.[202] Global antimicrobial resistance policies could take lessons from the environmental sector by adopting strategies that have made international environmental agreements successful in the past such as: sanctions for non-compliance, assistance for implementation, majority vote decision-making rules, an independent scientific panel, and specific commitments.[204]
United States
[edit]This section needs to be updated. (October 2023) |
For the United States 2016 budget, U.S. president Barack Obama proposed to nearly double the amount of federal funding to "combat and prevent" antibiotic resistance to more than $1.2 billion.[205] Many international funding agencies like USAID, DFID, SIDA and Bill & Melinda Gates Foundation have pledged money for developing strategies to counter antimicrobial resistance.[citation needed]
On 27 March 2015, the White House released a comprehensive plan to address the increasing need for agencies to combat the rise of antibiotic-resistant bacteria. The Task Force for Combating Antibiotic-Resistant Bacteria developed The National Action Plan for Combating Antibiotic-Resistant Bacteria with the intent of providing a roadmap to guide the US in the antibiotic resistance challenge and with hopes of saving many lives. This plan outlines steps taken by the Federal government over the next five years needed in order to prevent and contain outbreaks of antibiotic-resistant infections; maintain the efficacy of antibiotics already on the market; and to help to develop future diagnostics, antibiotics, and vaccines.[206]
The Action Plan was developed around five goals with focuses on strengthening health care, public health veterinary medicine, agriculture, food safety and research, and manufacturing. These goals, as listed by the White House, are as follows:
- Slow the Emergence of Resistant Bacteria and Prevent the Spread of Resistant Infections
- Strengthen National One-Health Surveillance Efforts to Combat Resistance
- Advance Development and use of Rapid and Innovative Diagnostic Tests for Identification and Characterization of Resistant Bacteria
- Accelerate Basic and Applied Research and Development for New Antibiotics, Other Therapeutics, and Vaccines
- Improve International Collaboration and Capacities for Antibiotic Resistance Prevention, Surveillance, Control and Antibiotic Research and Development
The following are goals set to meet by 2020:[206]
- Establishment of antimicrobial programs within acute care hospital settings
- Reduction of inappropriate antibiotic prescription and use by at least 50% in outpatient settings and 20% inpatient settings
- Establishment of State Antibiotic Resistance (AR) Prevention Programs in all 50 states
- Elimination of the use of medically important antibiotics for growth promotion in food-producing animals.
Current Status of AMR in the U.S.
As of 2023, antimicrobial resistance (AMR) remains a significant public health threat in the United States. According to the Centers for Disease Control and Prevention's 2023 Report on Antibiotic Resistance Threats, over 2.8 million antibiotic-resistant infections occur in the U.S. each year, leading to at least 35,000 deaths annually.[207] Among the most concerning resistant pathogens are Carbapenem-resistant Enterobacteriaceae (CRE), Methicillin-resistant Staphylococcus aureus (MRSA), and Clostridioides difficile (C. diff), all of which continue to be responsible for severe healthcare-associated infections (HAIs).
The COVID-19 pandemic led to a significant disruption in healthcare, with an increase in the use of antibiotics during the treatment of viral infections. This rise in antibiotic prescribing, coupled with overwhelmed healthcare systems, contributed to a resurgence in AMR during the pandemic years. A 2021 CDC report identified a sharp increase in HAIs caused by resistant pathogens in COVID-19 patients, a trend that has persisted into 2023.[208] Recent data suggest that although antibiotic use has decreased since the pandemic, some resistant pathogens remain prevalent in healthcare settings.[207]
The CDC has also expanded its Get Ahead of Sepsis campaign in 2023, focusing on raising awareness of AMR's role in sepsis and promoting the judicious use of antibiotics in both healthcare and community settings.[209] This initiative has reached millions through social media, healthcare facilities, and public health outreach, aiming to educate the public on the importance of preventing infections and reducing antibiotic misuse.
Policies
[edit]According to World Health Organization, policymakers can help tackle resistance by strengthening resistance-tracking and laboratory capacity and by regulating and promoting the appropriate use of medicines.[16] Policymakers and industry can help tackle resistance by: fostering innovation and research and development of new tools; and promoting cooperation and information sharing among all stakeholders.[16]
The U.S. government continues to prioritize AMR mitigation through policy and legislation. In 2023, the National Action Plan for Combating Antibiotic-Resistant Bacteria (CARB) 2023-2028 was released, outlining strategic objectives for reducing antibiotic-resistant infections, advancing infection prevention, and accelerating research on new antibiotics.[210] The plan also emphasizes the importance of improving antibiotic stewardship across healthcare, agriculture, and veterinary settings. Furthermore, the PASTEUR Act (Pioneering Antimicrobial Subscriptions to End Upsurging Resistance) has gained momentum in Congress. If passed, the bill would create a subscription-based payment model to incentivize the development of new antimicrobial drugs, while supporting antimicrobial stewardship programs to reduce the misuse of existing antibiotics.[211] This legislation is considered a critical step toward addressing the economic barriers to developing new antimicrobials.
Policy evaluation
[edit]Measuring the costs and benefits of strategies to combat AMR is difficult and policies may only have effects in the distant future. In other infectious diseases this problem has been addressed by using mathematical models. More research is needed to understand how AMR develops and spreads so that mathematical modelling can be used to anticipate the likely effects of different policies.[212]
Further research
[edit]Rapid testing and diagnostics
[edit]
Distinguishing infections requiring antibiotics from self-limiting ones is clinically challenging. In order to guide appropriate use of antibiotics and prevent the evolution and spread of antimicrobial resistance, diagnostic tests that provide clinicians with timely, actionable results are needed.
Acute febrile illness is a common reason for seeking medical care worldwide and a major cause of morbidity and mortality. In areas with decreasing malaria incidence, many febrile patients are inappropriately treated for malaria, and in the absence of a simple diagnostic test to identify alternative causes of fever, clinicians presume that a non-malarial febrile illness is most likely a bacterial infection, leading to inappropriate use of antibiotics. Multiple studies have shown that the use of malaria rapid diagnostic tests without reliable tools to distinguish other fever causes has resulted in increased antibiotic use.[213]
Antimicrobial susceptibility testing (AST) can facilitate a precision medicine approach to treatment by helping clinicians to prescribe more effective and targeted antimicrobial therapy.[214] At the same time with traditional phenotypic AST it can take 12 to 48 hours to obtain a result due to the time taken for organisms to grow on/in culture media.[215] Rapid testing, possible from molecular diagnostics innovations, is defined as "being feasible within an 8-h working shift".[215] There are several commercial Food and Drug Administration-approved assays available which can detect AMR genes from a variety of specimen types. Progress has been slow due to a range of reasons including cost and regulation.[216] Genotypic AMR characterisation methods are, however, being increasingly used in combination with machine learning algorithms in research to help better predict phenotypic AMR from organism genotype.[217][218]
Optical techniques such as phase contrast microscopy in combination with single-cell analysis are another powerful method to monitor bacterial growth. In 2017, scientists from Uppsala University in Sweden published a method[219] that applies principles of microfluidics and cell tracking, to monitor bacterial response to antibiotics in less than 30 minutes overall manipulation time. This invention was awarded the 8M£ Longitude Prize on AMR in 2024. Recently, this platform has been advanced by coupling microfluidic chip with optical tweezing[220] in order to isolate bacteria with altered phenotype directly from the analytical matrix.
Rapid diagnostic methods have also been trialled as antimicrobial stewardship interventions to influence the healthcare drivers of AMR. Serum procalcitonin measurement has been shown to reduce mortality rate, antimicrobial consumption and antimicrobial-related side-effects in patients with respiratory infections, but impact on AMR has not yet been demonstrated.[221] Similarly, point of care serum testing of the inflammatory biomarker C-reactive protein has been shown to influence antimicrobial prescribing rates in this patient cohort, but further research is required to demonstrate an effect on rates of AMR.[222] Clinical investigation to rule out bacterial infections are often done for patients with pediatric acute respiratory infections. Currently it is unclear if rapid viral testing affects antibiotic use in children.[223]
Vaccines
[edit]Vaccines are an essential part of the response to reduce AMR as they prevent infections, reduce the use and overuse of antimicrobials, and slow the emergence and spread of drug-resistant pathogens.[7] Microorganisms usually do not develop resistance to vaccines because vaccines reduce the spread of the infection and target the pathogen in multiple ways in the same host and possibly in different ways between different hosts. Furthermore, if the use of vaccines increases, there is evidence that antibiotic resistant strains of pathogens will decrease; the need for antibiotics will naturally decrease as vaccines prevent infection before it occurs.[224] A 2024 report by WHO finds that vaccines against 24 pathogens could reduce the number of antibiotics needed by 22% or 2.5 billion defined daily doses globally every year.[7] If vaccines could be rolled out against all the evaluated pathogens, they could save a third of the hospital costs associated with AMR.[7] Vaccinated people have fewer infections and are protected against potential complications from secondary infections that may need antimicrobial medicines or require admission to hospital.[7] However, there are well documented cases of vaccine resistance, although these are usually much less of a problem than antimicrobial resistance.[225][226]
While theoretically promising, antistaphylococcal vaccines have shown limited efficacy, because of immunological variation between Staphylococcus species, and the limited duration of effectiveness of the antibodies produced. Development and testing of more effective vaccines is underway.[227]
Two registrational trials have evaluated vaccine candidates in active immunization strategies against S. aureus infection. In a phase II trial, a bivalent vaccine of capsular proteins 5 & 8 was tested in 1804 hemodialysis patients with a primary fistula or synthetic graft vascular access. After 40 weeks following vaccination a protective effect was seen against S. aureus bacteremia, but not at 54 weeks following vaccination.[228] Based on these results, a second trial was conducted which failed to show efficacy.[229]
Merck tested V710, a vaccine targeting IsdB, in a blinded randomized trial in patients undergoing median sternotomy. The trial was terminated after a higher rate of multiorgan system failure–related deaths was found in the V710 recipients. Vaccine recipients who developed S. aureus infection were five times more likely to die than control recipients who developed S. aureus infection.[230]
Numerous investigators have suggested that a multiple-antigen vaccine would be more effective, but a lack of biomarkers defining human protective immunity keep these proposals in the logical, but strictly hypothetical arena.[229]
Antibody therapy
[edit]Antibodies are promising against antimicrobial resistance. Monoclonal antibodies (mAbs) target bacterial virulence factors, aiding in bacterial destruction through various mechanisms. Three FDA-approved antibodies target B. anthracis and C. difficile toxins.[231][232] Innovative strategies include DSTA4637S, an antibody-antibiotic conjugate, and MEDI13902, a bispecific antibody targeting Pseudomonas aeruginosa components.[232]
Alternating therapy
[edit]Alternating therapy is a proposed method in which two or three antibiotics are taken in a rotation versus taking just one antibiotic such that bacteria resistant to one antibiotic are killed when the next antibiotic is taken. Studies have found that this method reduces the rate at which antibiotic resistant bacteria emerge in vitro relative to a single drug for the entire duration.[233]
Studies have found that bacteria that evolve antibiotic resistance towards one group of antibiotic may become more sensitive to others.[234] This phenomenon can be used to select against resistant bacteria using an approach termed collateral sensitivity cycling, which has recently been found to be relevant in developing treatment strategies for chronic infections caused by Pseudomonas aeruginosa.[235] Despite its promise, large-scale clinical and experimental studies revealed limited evidence of susceptibility to antibiotic cycling across various pathogens.[236][237]
Development of new drugs
[edit]Since the discovery of antibiotics, research and development (R&D) efforts have provided new drugs in time to treat bacteria that became resistant to older antibiotics, but in the 2000s there has been concern that development has slowed enough that seriously ill people may run out of treatment options.[238][239] Another concern is that practitioners may become reluctant to perform routine surgeries because of the increased risk of harmful infection.[240] Backup treatments can have serious side-effects; for example, antibiotics like aminoglycosides (such as amikacin, gentamicin, kanamycin, streptomycin, etc.) used for the treatment of drug-resistant tuberculosis and cystic fibrosis can cause respiratory disorders, deafness and kidney failure.[241][242]
The potential crisis at hand is the result of a marked decrease in industry research and development.[243][244] Poor financial investment in antibiotic research has exacerbated the situation.[245][243] The pharmaceutical industry has little incentive to invest in antibiotics because of the high risk and because the potential financial returns are less likely to cover the cost of development than for other pharmaceuticals.[246] In 2011, Pfizer, one of the last major pharmaceutical companies developing new antibiotics, shut down its primary research effort, citing poor shareholder returns relative to drugs for chronic illnesses.[247] However, small and medium-sized pharmaceutical companies are still active in antibiotic drug research. In particular, apart from classical synthetic chemistry methodologies, researchers have developed a combinatorial synthetic biology platform on single cell level in a high-throughput screening manner to diversify novel lanthipeptides.[248]
In the 5–10 years since 2010, there has been a significant change in the ways new antimicrobial agents are discovered and developed – principally via the formation of public-private funding initiatives. These include CARB-X,[249] which focuses on nonclinical and early phase development of novel antibiotics, vaccines, rapid diagnostics; Novel Gram Negative Antibiotic (GNA-NOW),[250] which is part of the EU's Innovative Medicines Initiative; and Replenishing and Enabling the Pipeline for Anti-infective Resistance Impact Fund (REPAIR).[251] Later stage clinical development is supported by the AMR Action Fund, which in turn is supported by multiple investors with the aim of developing 2–4 new antimicrobial agents by 2030. The delivery of these trials is facilitated by national and international networks supported by the Clinical Research Network of the National Institute for Health and Care Research (NIHR), European Clinical Research Alliance in Infectious Diseases (ECRAID) and the recently formed ADVANCE-ID, which is a clinical research network based in Asia.[252] The Global Antibiotic Research and Development Partnership (GARDP) is generating new evidence for global AMR threats such as neonatal sepsis, treatment of serious bacterial infections and sexually transmitted infections as well as addressing global access to new and strategically important antibacterial drugs.[253]
The discovery and development of new antimicrobial agents has been facilitated by regulatory advances, which have been principally led by the European Medicines Agency (EMA) and the Food and Drug Administration (FDA). These processes are increasingly aligned although important differences remain and drug developers must prepare separate documents. New development pathways have been developed to help with the approval of new antimicrobial agents that address unmet needs such as the Limited Population Pathway for Antibacterial and Antifungal Drugs (LPAD). These new pathways are required because of difficulties in conducting large definitive phase III clinical trials in a timely way.
Some of the economic impediments to the development of new antimicrobial agents have been addressed by innovative reimbursement schemes that delink payment of antimicrobials from volume-based sales. In the UK, a market entry reward scheme has been pioneered by the National Institute for Clinical Excellence (NICE) whereby an annual subscription fee is paid for use of strategically valuable antimicrobial agents – cefiderocol and ceftazidime-aviabactam are the first agents to be used in this manner and the scheme is potential blueprint for comparable programs in other countries.
The available classes of antifungal drugs are still limited but as of 2021 novel classes of antifungals are being developed and are undergoing various stages of clinical trials to assess performance.[254]
Scientists have started using advanced computational approaches with supercomputers for the development of new antibiotic derivatives to deal with antimicrobial resistance.[244][255][256]
Biomaterials
[edit]Using antibiotic-free alternatives in bone infection treatment may help decrease the use of antibiotics and thus antimicrobial resistance.[36] The bone regeneration material bioactive glass S53P4 has shown to effectively inhibit the bacterial growth of up to 50 clinically relevant bacteria including MRSA and MRSE.[257][258][259]
Nanomaterials
[edit]During the last decades, copper and silver nanomaterials have demonstrated appealing features for the development of a new family of antimicrobial agents.[260] Nanoparticles (1-100 nm) show unique properties and promise as antimicrobial agents against resistant bacteria. Silver (AgNPs) and gold nanoparticles (AuNPs) are extensively studied, disrupting bacterial cell membranes and interfering with protein synthesis. Zinc oxide (ZnO NPs), copper (CuNPs), and silica (SiNPs) nanoparticles also exhibit antimicrobial properties. However, high synthesis costs, potential toxicity, and instability pose challenges. To overcome these, biological synthesis methods and combination therapies with other antimicrobials are explored. Enhanced biocompatibility and targeting are also under investigation to improve efficacy.[232]
Rediscovery of ancient treatments
[edit]Similar to the situation in malaria therapy, where successful treatments based on ancient recipes have been found,[261] there has already been some success in finding and testing ancient drugs and other treatments that are effective against AMR bacteria.[262]
Computational community surveillance
[edit]One of the key tools identified by the WHO and others for the fight against rising antimicrobial resistance is improved surveillance of the spread and movement of AMR genes through different communities and regions. Recent advances in high-throughput DNA sequencing as a result of the Human Genome Project have resulted in the ability to determine the individual microbial genes in a sample.[263] Along with the availability of databases of known antimicrobial resistance genes, such as the Comprehensive Antimicrobial Resistance Database (CARD)[264][265] and ResFinder,[266][267] this allows the identification of all the antimicrobial resistance genes within the sample – the so-called "resistome". In doing so, a profile of these genes within a community or environment can be determined, providing insights into how antimicrobial resistance is spreading through a population and allowing for the identification of resistance that is of concern.[263]
Phage therapy
[edit]Phage therapy is the therapeutic use of bacteriophages to treat pathogenic bacterial infections.[268] Phage therapy has many potential applications in human medicine as well as dentistry, veterinary science, and agriculture.[269]
Phage therapy relies on the use of naturally occurring bacteriophages to infect and lyse bacteria at the site of infection in a host. Due to current advances in genetics and biotechnology these bacteriophages can possibly be manufactured to treat specific infections.[270] Phages can be bioengineered to target multidrug-resistant bacterial infections, and their use involves the added benefit of preventing the elimination of beneficial bacteria in the human body.[31] Phages destroy bacterial cell walls and membrane through the use of lytic proteins which kill bacteria by making many holes from the inside out.[271] Bacteriophages can even possess the ability to digest the biofilm that many bacteria develop that protect them from antibiotics in order to effectively infect and kill bacteria. Bioengineering can play a role in creating successful bacteriophages.[271]
Understanding the mutual interactions and evolutions of bacterial and phage populations in the environment of a human or animal body is essential for rational phage therapy.[272]
Bacteriophagics are used against antibiotic resistant bacteria in Georgia (George Eliava Institute) and in one institute in Wrocław, Poland.[273][274] Bacteriophage cocktails are common drugs sold over the counter in pharmacies in eastern countries.[275][276] In Belgium, four patients with severe musculoskeletal infections received bacteriophage therapy with concomitant antibiotics. After a single course of phage therapy, no recurrence of infection occurred and no severe side-effects related to the therapy were detected.[277]
See also
[edit]- Alliance for the Prudent Use of Antibiotics
- Antimicrobial resistance in Australia
- Broad-spectrum antibiotic
- Colonisation resistance
- Drug of last resort
- Genetic engineering
- (KPC) antibacterial resistance gene
- Multidrug-resistant Gram-negative bacteria
- Multidrug-resistant tuberculosis
- New Delhi metallo-beta-lactamase 1
- Persister cells
- Resistance-nodulation-cell division superfamily (RND)
- Resistome
References
[edit]- ^ Kirby-Bauer Disk Diffusion Susceptibility Test Protocol Archived 26 June 2011 at the Wayback Machine, Jan Hudzicki, ASM
- ^ a b "About Antimicrobial Resistance". US Centers for Disease Control and Prevention. 22 April 2024. Retrieved 11 October 2024.
- ^ a b c "Antimicrobial resistance Fact sheet N°194". who.int. April 2014. Archived from the original on 10 March 2015. Retrieved 7 March 2015.
- ^ a b Tanwar J, Das S, Fatima Z, Hameed S (2014). "Multidrug resistance: an emerging crisis". Interdisciplinary Perspectives on Infectious Diseases. 2014: 541340. doi:10.1155/2014/541340. PMC 4124702. PMID 25140175.
- ^ Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. (March 2012). "Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance". Clinical Microbiology and Infection. 18 (3): 268–281. doi:10.1111/j.1469-0691.2011.03570.x. PMID 21793988.
- ^ a b c d Dabour R, Meirson T, Samson AO (December 2016). "Global antibiotic resistance is mostly periodic". Journal of Global Antimicrobial Resistance. 7: 132–134. doi:10.1016/j.jgar.2016.09.003. PMID 27788414.
- ^ a b c d e f g h i j "Better use of vaccines could reduce antibiotic use by 2.5 billion doses annually, says WHO". World Health Organization. 10 October 2024. Retrieved 11 October 2024.
- ^ a b Saha M, Sarkar A (December 2021). "Review on Multiple Facets of Drug Resistance: A Rising Challenge in the 21st Century". Journal of Xenobiotics. 11 (4): 197–214. doi:10.3390/jox11040013. PMC 8708150. PMID 34940513.
- ^ Swedish work on containment of antibiotic resistance – Tools, methods and experiences (PDF). Stockholm: Public Health Agency of Sweden. 2014. pp. 16–17, 121–128. ISBN 978-91-7603-011-0. Archived (PDF) from the original on 23 July 2015. Retrieved 23 July 2015.
- ^ "Antimicrobial resistance". www.who.int. Retrieved 20 November 2024.
- ^ Chanel S, Doherty B (10 September 2020). "'Superbugs' a far greater risk than Covid in Pacific, scientist warns". The Guardian. ISSN 0261-3077. Archived from the original on 5 December 2022. Retrieved 14 September 2020.
- ^ Rodríguez-Baño J, Rossolini GM, Schultsz C, Tacconelli E, Murthy S, Ohmagari N, et al. (March 2021). "Key considerations on the potential impacts of the COVID-19 pandemic on antimicrobial resistance research and surveillance". Trans R Soc Trop Med Hyg. 115 (10): 1122–1129. doi:10.1093/trstmh/trab048. PMC 8083707. PMID 33772597.
- ^ "What is Drug Resistance?". niaid.nih.gov. Archived from the original on 27 July 2015. Retrieved 26 July 2015.
- ^ "CDC: Get Smart: Know When Antibiotics Work". Cdc.gov. 29 May 2018. Archived from the original on 29 April 2015. Retrieved 12 June 2013.
- ^ MacGowan A, Macnaughton E (1 October 2017). "Antibiotic resistance". Medicine. 45 (10): 622–628. doi:10.1016/j.mpmed.2017.07.006.
- ^ a b c d e "WHO's first global report on antibiotic resistance reveals serious, worldwide threat to public health" Archived 2 May 2014 at the Wayback Machine Retrieved 2 May 2014
- ^ a b c Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, et al. (Antimicrobial Resistance Collaborators) (February 2022). "Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis". Lancet. 399 (10325): 629–655. doi:10.1016/S0140-6736(21)02724-0. PMC 8841637. PMID 35065702. S2CID 246077406.
- ^ a b "Antibiotic resistance". who.int. Archived from the original on 21 May 2021. Retrieved 16 March 2020.
- ^ Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, et al. (January 2019). "Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis". The Lancet. Infectious Diseases. 19 (1): 56–66. doi:10.1016/S1473-3099(18)30605-4. PMC 6300481. PMID 30409683.
- ^ "Antibiotic-resistant bacteria responsible for over 33,000 deaths in Europe in 2015, study finds". The Pharmaceutical Journal. 7 November 2018. Archived from the original on 28 March 2023. Retrieved 28 March 2023.
- ^ Mestrovic T, Robles Aguilar G, Swetschinski LR, Ikuta KS, Gray AP, Davis Weaver N, et al. (November 2022). "The burden of bacterial antimicrobial resistance in the WHO European region in 2019: a cross-country systematic analysis". The Lancet. Public Health. 7 (11): e897 – e913. doi:10.1016/S2468-2667(22)00225-0. hdl:10023/26218. PMC 9630253. PMID 36244350.
- ^ "Antimicrobial Resistance " Cambridge Medicine Journal". Retrieved 27 February 2020.[permanent dead link]
- ^ a b Holmes AH, Moore LS, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, et al. (January 2016). "Understanding the mechanisms and drivers of antimicrobial resistance". Lancet. 387 (10014): 176–87. doi:10.1016/S0140-6736(15)00473-0. hdl:10044/1/32225. PMID 26603922. S2CID 1944665. Archived from the original on 14 April 2022. Retrieved 5 December 2021.
- ^ a b "CDC Newsroom". CDC. 1 January 2016. Archived from the original on 9 March 2023. Retrieved 28 February 2023.
- ^ a b Michael CA, Dominey-Howes D, Labbate M (2014). "The antimicrobial resistance crisis: causes, consequences, and management". Frontiers in Public Health. 2: 145. doi:10.3389/fpubh.2014.00145. PMC 4165128. PMID 25279369.
- ^ a b "Natural selection". evolution.berkeley.edu. Archived from the original on 30 October 2019. Retrieved 10 March 2020.
- ^ a b Larsen J, Raisen CL, Ba X, Sadgrove NJ, Padilla-González GF, Simmonds MS, et al. (February 2022). "Emergence of methicillin resistance predates the clinical use of antibiotics". Nature. 602 (7895): 135–141. Bibcode:2022Natur.602..135L. doi:10.1038/s41586-021-04265-w. PMC 8810379. PMID 34987223.
- ^ Crits-Christoph A, Hallowell HA, Koutouvalis K, Suez J (31 December 2022). "Good microbes, bad genes? The dissemination of antimicrobial resistance in the human microbiome". Gut Microbes. 14 (1): 2055944. doi:10.1080/19490976.2022.2055944. PMC 8959533. PMID 35332832.
- ^ a b c Ferri M, Ranucci E, Romagnoli P, Giaccone V (September 2017). "Antimicrobial resistance: A global emerging threat to public health systems". Critical Reviews in Food Science and Nutrition. 57 (13): 2857–2876. doi:10.1080/10408398.2015.1077192. PMID 26464037. S2CID 24549694.
- ^ "Global Database for Tracking Antimicrobial Resistance (AMR) Country Self- Assessment Survey (TrACSS)". amrcountryprogress.org. Archived from the original on 28 March 2023. Retrieved 28 March 2023.
- ^ a b c d Rather IA, Kim BC, Bajpai VK, Park YH (May 2017). "Self-medication and antibiotic resistance: Crisis, current challenges, and prevention". Saudi Journal of Biological Sciences. 24 (4): 808–812. doi:10.1016/j.sjbs.2017.01.004. PMC 5415144. PMID 28490950.
- ^ a b Torres NF, Chibi B, Middleton LE, Solomon VP, Mashamba-Thompson TP (March 2019). "Evidence of factors influencing self-medication with antibiotics in low and middle-income countries: a systematic scoping review". Public Health. 168: 92–101. doi:10.1016/j.puhe.2018.11.018. PMID 30716570. S2CID 73434085.
- ^ Ayukekbong JA, Ntemgwa M, Atabe AN (15 May 2017). "The threat of antimicrobial resistance in developing countries: causes and control strategies". Antimicrobial Resistance and Infection Control. 6 (1): 47. doi:10.1186/s13756-017-0208-x. PMC 5433038. PMID 28515903.
- ^ Chen C, Patterson B, Simpson R, Li Y, Chen Z, Lv Q, et al. (9 August 2022). "Do fluoroquinolones increase aortic aneurysm or dissection incidence and mortality? A systematic review and meta-analysis". Frontiers in Cardiovascular Medicine. 9: 949538. doi:10.3389/fcvm.2022.949538. PMC 9396038. PMID 36017083.
- ^ Shu Y, Zhang Q, He X, Liu Y, Wu P, Chen L (6 September 2022). "Fluoroquinolone-associated suspected tendonitis and tendon rupture: A pharmacovigilance analysis from 2016 to 2021 based on the FAERS database". Frontiers in Pharmacology. 13: 990241. doi:10.3389/fphar.2022.990241. PMC 9486157. PMID 36147351.
- ^ a b c Ventola CL (April 2015). "The antibiotic resistance crisis: part 1: causes and threats". P & T. 40 (4): 277–83. PMC 4378521. PMID 25859123.
- ^ Fleming-Dutra KE, Hersh AL, Shapiro DJ, Bartoces M, Enns EA, File TM, et al. (May 2016). "Prevalence of Inappropriate Antibiotic Prescriptions Among US Ambulatory Care Visits, 2010–2011". JAMA. 315 (17): 1864–73. doi:10.1001/jama.2016.4151. PMID 27139059.
- ^ Strachan CR, Davies J (February 2017). "The Whys and Wherefores of Antibiotic Resistance". Cold Spring Harbor Perspectives in Medicine. 7 (2): a025171. doi:10.1101/cshperspect.a025171. PMC 5287056. PMID 27793964.
- ^ Harris A, Chandramohan S, Awali RA, Grewal M, Tillotson G, Chopra T (August 2019). "Physicians' attitude and knowledge regarding antibiotic use and resistance in ambulatory settings". American Journal of Infection Control. 47 (8): 864–868. doi:10.1016/j.ajic.2019.02.009. PMID 30926215. S2CID 88482220.
- ^ Joshi MP (17 February 2021). "Don't let Covid boost another killer". Knowable Magazine. doi:10.1146/knowable-021621-1. Archived from the original on 22 October 2021. Retrieved 10 August 2022.
- ^ Rawson TM, Moore LS, Zhu N, Ranganathan N, Skolimowska K, Gilchrist M, et al. (December 2020). "Bacterial and Fungal Coinfection in Individuals With Coronavirus: A Rapid Review To Support COVID-19 Antimicrobial Prescribing". Clinical Infectious Diseases. 71 (9): 2459–2468. doi:10.1093/cid/ciaa530. PMC 7197596. PMID 32358954.
- ^ Barnes S. "Rutgers study finds antibiotic overuse is caused by misconceptions, financial incentives". The Daily Targum. Archived from the original on 6 December 2021. Retrieved 16 February 2021.
- ^ Blaser MJ, Melby MK, Lock M, Nichter M (February 2021). "Accounting for variation in and overuse of antibiotics among humans". BioEssays. 43 (2): e2000163. doi:10.1002/bies.202000163. PMID 33410142. S2CID 230811912.
- ^ "Antimicrobials | American Veterinary Medical Association". avma.org. Archived from the original on 24 April 2024. Retrieved 24 April 2024.
- ^ Caneschi A, Bardhi A, Barbarossa A, Zaghini A (March 2023). "The Use of Antibiotics and Antimicrobial Resistance in Veterinary Medicine, a Complex Phenomenon: A Narrative Review". Antibiotics. 12 (3): 487. doi:10.3390/antibiotics12030487. PMC 10044628. PMID 36978354.
- ^ "Has COVID-19 made the superbug crisis worse?". Global News. Archived from the original on 12 February 2022. Retrieved 12 February 2022.
- ^ Lucien MA, Canarie MF, Kilgore PE, Jean-Denis G, Fénélon N, Pierre M, et al. (March 2021). "Antibiotics and antimicrobial resistance in the COVID-19 era: Perspective from resource-limited settings". International Journal of Infectious Diseases. 104: 250–254. doi:10.1016/j.ijid.2020.12.087. PMC 7796801. PMID 33434666.
- ^ "COVID-19 & Antibiotic Resistance". Centers for Disease Control and Prevention. 18 November 2021. Archived from the original on 21 February 2022. Retrieved 21 February 2022.
- ^ Knight GM, Glover RE, McQuaid CF, Olaru ID, Gallandat K, Leclerc QJ, et al. (February 2021). "Antimicrobial resistance and COVID-19: Intersections and implications". eLife. 10. doi:10.7554/eLife.64139. PMC 7886324. PMID 33588991. S2CID 231936902.
- ^ Lu J, Guo J (January 2021). "Disinfection spreads antimicrobial resistance". Science. 371 (6528): 474. Bibcode:2021Sci...371..474L. doi:10.1126/science.abg4380. PMID 33510019. S2CID 231730007.
- ^ Lobie TA, Roba AA, Booth JA, Kristiansen KI, Aseffa A, Skarstad K, et al. (October 2021). "Antimicrobial resistance: A challenge awaiting the post-COVID-19 era". International Journal of Infectious Diseases. 111: 322–325. doi:10.1016/j.ijid.2021.09.003. PMC 8425743. PMID 34508864. S2CID 237444117.
- ^ "World Leaders Approve Milestone Commitment To Reduce Deaths From Antibiotic Resistance By 10% By 2030 - Health Policy Watch". 26 September 2024. Retrieved 28 September 2024.
- ^ "UN General Assembly High-Level Meeting on antimicrobial resistance 2024". www.who.int. Retrieved 28 September 2024.
- ^ Musoke D, Namata C, Lubega GB, Niyongabo F, Gonza J, Chidziwisano K, et al. (October 2021). "The role of Environmental Health in preventing antimicrobial resistance in low- and middle-income countries". Environmental Health and Preventive Medicine. 26 (1): 100. Bibcode:2021EHPM...26..100M. doi:10.1186/s12199-021-01023-2. PMC 8493696. PMID 34610785.
- ^ a b c d e Fletcher S (July 2015). "Understanding the contribution of environmental factors in the spread of antimicrobial resistance". Environmental Health and Preventive Medicine. 20 (4): 243–252. Bibcode:2015EHPM...20..243F. doi:10.1007/s12199-015-0468-0. PMC 4491066. PMID 25921603.
- ^ a b c d e f Ahmad I, Malak HA, Abulreesh HH (December 2021). "Environmental antimicrobial resistance and its drivers: a potential threat to public health". Journal of Global Antimicrobial Resistance. 27: 101–111. doi:10.1016/j.jgar.2021.08.001. PMID 34454098.
- ^ Monger XC, Gilbert AA, Saucier L, Vincent AT (October 2021). "Antibiotic Resistance: From Pig to Meat". Antibiotics. 10 (10): 1209. doi:10.3390/antibiotics10101209. PMC 8532907. PMID 34680790.
- ^ Torrella K (8 January 2023). "Big Meat just can't quit antibiotics". Vox. Archived from the original on 23 January 2023. Retrieved 23 January 2023.
- ^ a b Martin MJ, Thottathil SE, Newman TB (December 2015). "Antibiotics Overuse in Animal Agriculture: A Call to Action for Health Care Providers". American Journal of Public Health. 105 (12): 2409–2410. doi:10.2105/AJPH.2015.302870. PMC 4638249. PMID 26469675.
- ^ "Farm antibiotic use". saveourantibiotics.org. Archived from the original on 3 April 2024. Retrieved 21 March 2024.
- ^ Tang KL, Caffrey NP, Nóbrega DB, Cork SC, Ronksley PE, Barkema HW, et al. (November 2017). "Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis". The Lancet. Planetary Health. 1 (8): e316 – e327. doi:10.1016/S2542-5196(17)30141-9. PMC 5785333. PMID 29387833.
- ^ a b Innes GK, Randad PR, Korinek A, Davis MF, Price LB, So AD, et al. (April 2020). "External Societal Costs of Antimicrobial Resistance in Humans Attributable to Antimicrobial Use in Livestock". Annual Review of Public Health. 41 (1): 141–157. doi:10.1146/annurev-publhealth-040218-043954. PMC 7199423. PMID 31910712.
- ^ "Veterinary feed directive (VFD) basics | American Veterinary Medical Association". avma.org. Archived from the original on 24 April 2024. Retrieved 24 April 2024.
- ^ US EPA O (15 March 2013). "What are Antimicrobial Pesticides?". US EPA. Archived from the original on 27 November 2022. Retrieved 28 February 2020.
- ^ Ramakrishnan B, Venkateswarlu K, Sethunathan N, Megharaj M (March 2019). "Local applications but global implications: Can pesticides drive microorganisms to develop antimicrobial resistance?". The Science of the Total Environment. 654: 177–189. Bibcode:2019ScTEn.654..177R. doi:10.1016/j.scitotenv.2018.11.041. PMID 30445319. S2CID 53568193.
- ^ Rhodes J, Abdolrasouli A, Dunne K, Sewell TR, Zhang Y, Ballard E, et al. (May 2022). "Population genomics confirms acquisition of drug-resistant Aspergillus fumigatus infection by humans from the environment". Nature Microbiology. 7 (5): 663–674. doi:10.1038/s41564-022-01091-2. PMC 9064804. PMID 35469019.
- ^ a b Verweij PE, Arendrup MC, Alastruey-Izquierdo A, Gold JA, Lockhart SR, Chiller T, et al. (December 2022). "Dual use of antifungals in medicine and agriculture: How do we help prevent resistance developing in human pathogens?". Drug Resistance Updates. 65: 100885. doi:10.1016/j.drup.2022.100885. PMC 10693676. PMID 36283187. S2CID 253052170.
- ^ a b Perron GG, Whyte L, Turnbaugh PJ, Goordial J, Hanage WP, Dantas G, et al. (25 March 2015). "Functional characterization of bacteria isolated from ancient arctic soil exposes diverse resistance mechanisms to modern antibiotics". PLOS ONE. 10 (3): e0069533. Bibcode:2015PLoSO..1069533P. doi:10.1371/journal.pone.0069533. PMC 4373940. PMID 25807523.
- ^ McGee D, Gribkoff E (4 August 2022). "Permafrost". MIT Climate Portal. Archived from the original on 27 September 2023. Retrieved 27 September 2023.
- ^ Fox-Kemper, B., H.T. Hewitt, C. Xiao, G. Aðalgeirsdóttir, S.S. Drijfhout, T.L. Edwards, N.R. Golledge, M. Hemer, R.E. Kopp, G. Krinner, A. Mix, D. Notz, S. Nowicki, I.S. Nurhati, L. Ruiz, J.-B. Sallée, A.B.A. Slangen, and Y. Yu, 2021: Chapter 9: Ocean, Cryosphere and Sea Level Change Archived 24 October 2022 at the Wayback Machine. In Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change [Masson-Delmotte, V., P. Zhai, A. Pirani, S.L. Connors, C. Péan, S. Berger, N. Caud, Y. Chen, L. Goldfarb, M.I. Gomis, M. Huang, K. Leitzell, E. Lonnoy, J.B.R. Matthews, T.K. Maycock, T. Waterfield, O. Yelekçi, R. Yu, and B. Zhou (eds.)]. Cambridge University Press, Cambridge, United Kingdom and New York, NY, USA, pp. 1211–1362, doi:10.1017/9781009157896.011.
- ^ Michaeleen D. "Are There Zombie Viruses — Like The 1918 Flu — Thawing In The Permafrost?". NPR.org. Archived from the original on 24 April 2023. Retrieved 4 April 2023.
- ^ "Anthrax Outbreak In Russia Thought To Be Result Of Thawing Permafrost". NPR.org. Archived from the original on 22 September 2016. Retrieved 24 September 2016.
- ^ Ng TF, Chen LF, Zhou Y, Shapiro B, Stiller M, Heintzman PD, et al. (November 2014). "Preservation of viral genomes in 700-y-old caribou feces from a subarctic ice patch". Proceedings of the National Academy of Sciences of the United States of America. 111 (47): 16842–16847. Bibcode:2014PNAS..11116842N. doi:10.1073/pnas.1410429111. PMC 4250163. PMID 25349412.
- ^ Legendre M, Lartigue A, Bertaux L, Jeudy S, Bartoli J, Lescot M, et al. (September 2015). "In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba". Proceedings of the National Academy of Sciences of the United States of America. 112 (38): E5327 – E5335. Bibcode:2015PNAS..112E5327L. doi:10.1073/pnas.1510795112. JSTOR 26465169. PMC 4586845. PMID 26351664.
- ^ Alempic JM, Lartigue A, Goncharov AE, Grosse G, Strauss J, Tikhonov AN, et al. (February 2023). "An Update on Eukaryotic Viruses Revived from Ancient Permafrost". Viruses. 15 (2): 564. doi:10.3390/v15020564. PMC 9958942. PMID 36851778.
- ^ Alund NN (9 March 2023). "Scientists revive 'zombie virus' that was frozen for nearly 50,000 years". USA Today. Archived from the original on 24 April 2023. Retrieved 23 April 2023.
- ^ Sajjad W, Rafiq M, Din G, Hasan F, Iqbal A, Zada S, et al. (September 2020). "Resurrection of inactive microbes and resistome present in the natural frozen world: Reality or myth?". The Science of the Total Environment. 735: 139275. Bibcode:2020ScTEn.73539275S. doi:10.1016/j.scitotenv.2020.139275. PMID 32480145. S2CID 219169932.
- ^ Miner KR, D'Andrilli J, Mackelprang R, Edwards A, Malaska MJ, Waldrop MP, et al. (30 September 2021). "Emergent biogeochemical risks from Arctic permafrost degradation". Nature Climate Change. 11 (1): 809–819. Bibcode:2021NatCC..11..809M. doi:10.1038/s41558-021-01162-y. S2CID 238234156.
- ^ Wu R, Trubl G, Taş N, Jansson JK (15 April 2022). "Permafrost as a potential pathogen reservoir". One Earth. 5 (4): 351–360. Bibcode:2022OEart...5..351W. doi:10.1016/j.oneear.2022.03.010. S2CID 248208195.
- ^ Rogers Van Katwyk S, Giubilini A, Kirchhelle C, Weldon I, Harrison M, McLean A, et al. (March 2023). "Exploring Models for an International Legal Agreement on the Global Antimicrobial Commons: Lessons from Climate Agreements". Health Care Analysis. 31 (1): 25–46. doi:10.1007/s10728-019-00389-3. PMC 10042908. PMID 31965398.
- ^ Wilson LA, Van Katwyk SR, Weldon I, Hoffman SJ (2021). "A Global Pandemic Treaty Must Address Antimicrobial Resistance". The Journal of Law, Medicine & Ethics. 49 (4): 688–691. doi:10.1017/jme.2021.94. PMC 8749967. PMID 35006051.
- ^ a b c Spurling GK, Dooley L, Clark J, Askew DA, et al. (Cochrane Acute Respiratory Infections Group) (October 2023). "Immediate versus delayed versus no antibiotics for respiratory infections". The Cochrane Database of Systematic Reviews. 2023 (10): CD004417. doi:10.1002/14651858.CD004417.pub6. PMC 10548498. PMID 37791590.
- ^ a b c d "Duration of antibiotic therapy and resistance". NPS Medicinewise. National Prescribing Service Limited trading, Australia. 13 June 2013. Archived from the original on 23 July 2015. Retrieved 22 July 2015.
- ^ Abdelsalam Elshenawy R, Umaru N, Aslanpour Z (March 2024). "Shorter and Longer Antibiotic Durations for Respiratory Infections: To Fight Antimicrobial Resistance-A Retrospective Cross-Sectional Study in a Secondary Care Setting in the UK". Pharmaceuticals. 17 (3): 339. doi:10.3390/ph17030339. PMC 10975983. PMID 38543125.
- ^ a b c "Biggest Threats – Antibiotic/Antimicrobial Resistance – CDC". cdc.gov. 10 September 2018. Archived from the original on 12 September 2017. Retrieved 5 May 2016.
- ^ "HealthMap Resistance". HealthMap.org Boston Children's Hospital. Archived from the original on 15 November 2017. Retrieved 15 November 2017.
- ^ Scales D. "Mapping Antibiotic Resistance: Know The Germs in Your Neighborhood". Commonhealth. National Public Radio. Archived from the original on 8 December 2015. Retrieved 8 December 2015.
- ^ "ResistanceMap". Center for Disease Dynamics, Economics & Policy. Archived from the original on 14 November 2017. Retrieved 14 November 2017.
- ^ a b "Global Action Plan on Antimicrobial Resistance" (PDF). WHO. Archived from the original (PDF) on 31 October 2017. Retrieved 14 November 2017.
- ^ Mader R, Damborg P, Amat JP, Bengtsson B, Bourély C, Broens EM, et al. (January 2021). "Building the European Antimicrobial Resistance Surveillance network in veterinary medicine (EARS-Vet)". Euro Surveillance. 26 (4). doi:10.2807/1560-7917.ES.2021.26.4.2001359. PMC 7848785. PMID 33509339.
- ^ Feuer L, Frenzer SK, Merle R, Leistner R, Bäumer W, Bethe A, et al. (September 2024). "Prevalence of MRSA in canine and feline clinical samples from one-third of veterinary practices in Germany from 2019-2021". The Journal of Antimicrobial Chemotherapy. 79 (9): 2273–2280. doi:10.1093/jac/dkae225. PMID 39007221.
- ^ a b c d e f g Fisher MC, Alastruey-Izquierdo A, Berman J, Bicanic T, Bignell EM, Bowyer P, et al. (September 2022). "Tackling the emerging threat of antifungal resistance to human health". Nature Reviews. Microbiology. 20 (9): 557–571. doi:10.1038/s41579-022-00720-1. PMC 8962932. PMID 35352028.
- ^ Baur D, Gladstone BP, Burkert F, Carrara E, Foschi F, Döbele S, et al. (September 2017). "Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: a systematic review and meta-analysis". The Lancet. Infectious Diseases. 17 (9): 990–1001. doi:10.1016/S1473-3099(17)30325-0. PMID 28629876.
- ^ Gallagher JC, Justo JA, Chahine EB, Bookstaver PB, Scheetz M, Suda KJ, et al. (August 2018). "Preventing the Post-Antibiotic Era by Training Future Pharmacists as Antimicrobial Stewards". American Journal of Pharmaceutical Education. 82 (6): 6770. doi:10.5688/ajpe6770. PMC 6116871. PMID 30181677.
- ^ Andersson DI, Hughes D (September 2011). "Persistence of antibiotic resistance in bacterial populations". FEMS Microbiology Reviews. 35 (5): 901–11. doi:10.1111/j.1574-6976.2011.00289.x. PMID 21707669.
- ^ Gilberg K, Laouri M, Wade S, Isonaka S (2003). "Analysis of medication use patterns:apparent overuse of antibiotics and underuse of prescription drugs for asthma, depression, and CHF". Journal of Managed Care Pharmacy. 9 (3): 232–7. doi:10.18553/jmcp.2003.9.3.232. PMC 10437266. PMID 14613466. S2CID 25457069.
- ^ Llor C, Bjerrum L (December 2014). "Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem". Therapeutic Advances in Drug Safety. 5 (6): 229–41. doi:10.1177/2042098614554919. PMC 4232501. PMID 25436105.
- ^ "Pandemic of Antibiotic Resistance Killing Children in Bangladesh". Science Trend. 18 July 2021. Archived from the original on 29 November 2021. Retrieved 15 August 2021.
- ^ Chisti MJ, Harris JB, Carroll RW, Shahunja KM, Shahid AS, Moschovis PP, et al. (July 2021). "Antibiotic-Resistant Bacteremia in Young Children Hospitalized With Pneumonia in Bangladesh Is Associated With a High Mortality Rate". Open Forum Infectious Diseases. 8 (7): ofab260. doi:10.1093/ofid/ofab260. PMC 8280371. PMID 34277885.
- ^ The WHO AWaRe (Access, Watch, Reserve) antibiotic book. Geneva: World Health Organization (WHO). 2022. ISBN 978-92-4-006238-2. Archived from the original on 13 August 2023. Retrieved 28 March 2023.
- ^ Talento AF, Qualie M, Cottom L, Backx M, White PL (September 2021). "Lessons from an Educational Invasive Fungal Disease Conference on Hospital Antifungal Stewardship Practices across the UK and Ireland". Journal of Fungi. 7 (10): 801. doi:10.3390/jof7100801. PMC 8538376. PMID 34682223.
- ^ Doron S, Davidson LE (November 2011). "Antimicrobial stewardship". Mayo Clinic Proceedings. 86 (11): 1113–23. doi:10.4065/mcp.2011.0358. PMC 3203003. PMID 22033257.
- ^ Davey P, Marwick CA, Scott CL, Charani E, McNeil K, Brown E, et al. (February 2017). "Interventions to improve antibiotic prescribing practices for hospital inpatients". The Cochrane Database of Systematic Reviews. 2017 (2): CD003543. doi:10.1002/14651858.cd003543.pub4. PMC 6464541. PMID 28178770.
- ^ a b Costa T, Pimentel AC, Mota-Vieira L, Castanha AC (1 May 2021). "The benefits of a unit dose system in oral antibiotics dispensing: Azorean hospital pharmacists tackling the socioeconomic problem of leftovers in Portugal". Drugs & Therapy Perspectives. 37 (5): 212–221. doi:10.1007/s40267-021-00825-2. ISSN 1179-1977.
- ^ O'Sullivan JW, Harvey RT, Glasziou PP, McCullough A (November 2016). "Written information for patients (or parents of child patients) to reduce the use of antibiotics for acute upper respiratory tract infections in primary care". The Cochrane Database of Systematic Reviews. 2016 (11): CD011360. doi:10.1002/14651858.CD011360.pub2. PMC 6464519. PMID 27886368.
- ^ Bosley H, Henshall C, Appleton JV, Jackson D (March 2018). "A systematic review to explore influences on parental attitudes towards antibiotic prescribing in children" (PDF). Journal of Clinical Nursing. 27 (5–6): 892–905. doi:10.1111/jocn.14073. PMID 28906047. S2CID 4336064. Archived (PDF) from the original on 14 October 2023. Retrieved 6 May 2023.
- ^ "The Five Rights of Medication Administration". ihi.org. March 2007. Archived from the original on 24 October 2015. Retrieved 30 October 2015.
- ^ "CDC Features – Mission Critical: Preventing Antibiotic Resistance". cdc.gov. 4 April 2018. Archived from the original on 8 November 2017. Retrieved 22 July 2015.
- ^ Leekha S, Terrell CL, Edson RS (February 2011). "General principles of antimicrobial therapy". Mayo Clinic Proceedings. 86 (2): 156–67. doi:10.4065/mcp.2010.0639. PMC 3031442. PMID 21282489. Archived from the original on 15 September 2019. Retrieved 22 July 2015.
- ^ a b "Are you using antibiotics wisely?". Centers for Disease Control and Prevention. 3 January 2022. Archived from the original on 21 March 2024. Retrieved 21 March 2024.
- ^ "Articles". Cedars-Sinai. Archived from the original on 30 May 2020. Retrieved 23 March 2024.
- ^ "Defined Daily Dose (DDD)". WHO. Archived from the original on 10 May 2023. Retrieved 28 March 2023.
- ^ "Indicator: Antibiotic prescribing". QualityWatch. Nuffield Trust & Health Foundation. Archived from the original on 14 January 2015. Retrieved 16 July 2015.
- ^ a b c IACG (2018) Reduce unintentional exposure and the need for antimicrobials, and optimize their use IACG Discussion Paper Archived 6 July 2021 at the Wayback Machine, Interagency Coordination Group on Antimicrobial Resistance, public consultation process at WHO, Geneva, Switzerland
- ^ a b c d e Araya P (May 2016). "The Impact of Water and Sanitation on Diarrhoeal Disease Burden and Over-Consumption of Anitbiotics" (PDF). Archived (PDF) from the original on 1 October 2017. Retrieved 12 November 2017.
- ^ "Goal 6: Ensure availability and sustainable management of water and sanitation for all". United Nations Department of Economic and Social Affairs. Archived from the original on 24 September 2020. Retrieved 17 April 2023.
- ^ Swoboda SM, Earsing K, Strauss K, Lane S, Lipsett PA (February 2004). "Electronic monitoring and voice prompts improve hand hygiene and decrease nosocomial infections in an intermediate care unit". Critical Care Medicine. 32 (2): 358–63. doi:10.1097/01.CCM.0000108866.48795.0F. PMID 14758148. S2CID 9817602.(subscription required)
- ^ WHO, UNICEF (2015). Water, sanitation and hygiene in health care facilities – Status in low and middle income countries and way forward Archived 12 September 2018 at the Wayback Machine. World Health Organization (WHO), Geneva, Switzerland, ISBN 978 92 4 150847 6
- ^ Agga GE, Schmidt JW, Arthur TM (December 2016). "Effects of In-Feed Chlortetracycline Prophylaxis in Beef Cattle on Animal Health and Antimicrobial-Resistant Escherichia coli". Applied and Environmental Microbiology. 82 (24): 7197–7204. Bibcode:2016ApEnM..82.7197A. doi:10.1128/AEM.01928-16. PMC 5118930. PMID 27736789.
- ^ a b Brown EE, Cooper A, Carrillo C, Blais B (2019). "Selection of Multidrug-Resistant Bacteria in Medicated Animal Feeds". Frontiers in Microbiology. 10: 456. doi:10.3389/fmicb.2019.00456. PMC 6414793. PMID 30894847.
- ^ Marshall BM, Levy SB (October 2011). "Food animals and antimicrobials: impacts on human health". Clinical Microbiology Reviews. 24 (4): 718–33. doi:10.1128/CMR.00002-11. PMC 3194830. PMID 21976606.
- ^ Casewell M, Friis C, Marco E, McMullin P, Phillips I (August 2003). "The European ban on growth-promoting antibiotics and emerging consequences for human and animal health". The Journal of Antimicrobial Chemotherapy. 52 (2): 159–61. doi:10.1093/jac/dkg313. PMID 12837737.
- ^ Castanon JI (November 2007). "History of the use of antibiotic as growth promoters in European poultry feeds". Poultry Science. 86 (11): 2466–71. doi:10.3382/ps.2007-00249. PMID 17954599.(subscription required)
- ^ Bengtsson B, Wierup M (2006). "Antimicrobial resistance in Scandinavia after ban of antimicrobial growth promoters". Animal Biotechnology. 17 (2): 147–56. doi:10.1080/10495390600956920. PMID 17127526. S2CID 34602891.(subscription required)
- ^ Angulo FJ, Baker NL, Olsen SJ, Anderson A, Barrett TJ (April 2004). "Antimicrobial use in agriculture: controlling the transfer of antimicrobial resistance to humans". Seminars in Pediatric Infectious Diseases. 15 (2): 78–85. doi:10.1053/j.spid.2004.01.010. PMID 15185190.
- ^ "GAO-11-801, Antibiotic Resistance: Agencies Have Made Limited Progress Addressing Antibiotic Use in Animals". gao.gov. Archived from the original on 5 November 2013. Retrieved 25 January 2014.
- ^ Nelson JM, Chiller TM, Powers JH, Angulo FJ (April 2007). "Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story". Clinical Infectious Diseases. 44 (7): 977–80. doi:10.1086/512369. PMID 17342653.
- ^ "Extralabel Use and Antimicrobials". FDA. 29 April 2022. Archived from the original on 19 April 2023. Retrieved 19 April 2023.
- ^ Pallo-Zimmerman LM, Byron JK, Graves TK (July 2010). "Fluoroquinolones: then and now" (PDF). Compendium. 32 (7): E1-9, quiz E9. PMID 20957609. Archived (PDF) from the original on 21 June 2023. Retrieved 19 April 2023.
- ^ "RAND Europe Focus on Antimicrobial Resistance (AMR)". rand.org. Archived from the original on 21 April 2018. Retrieved 23 April 2018.
- ^ "React". Archived from the original on 16 November 2017. Retrieved 16 November 2017.
- ^ "Antibiotic Resistance: the silent tsunami (youtube video)". ReActTube. 6 March 2017. Archived from the original on 11 November 2021. Retrieved 17 November 2017.
- ^ "The Antibiotic Apocalypse Explained". Kurzgesagt – In a Nutshell. 16 March 2016. Archived from the original on 20 March 2021. Retrieved 17 November 2017.
- ^ "Ireland's National Action Plan on Antimicrobial Resistance 2017 – 2020". October 2017. Archived from the original on 10 August 2022. Retrieved 11 January 2019 – via Lenus (Irish Health Repository).
- ^ SARI Hospital Antimicrobial Stewardship Working Group (2009). Guidelines for antimicrobial stewardship in hospitals in Ireland. Dublin: HSE Health Protection Surveillance Centre (HPSC). ISBN 978-0-9551236-7-2. Archived from the original on 5 December 2021. Retrieved 11 January 2019.
- ^ "Taking antibiotics for colds and flu? There's no point". HSE.ie. Archived from the original on 28 May 2018. Retrieved 11 January 2019.
- ^ Murphy M, Bradley CP, Byrne S (May 2012). "Antibiotic prescribing in primary care, adherence to guidelines and unnecessary prescribing—an Irish perspective". BMC Family Practice. 13 (1): 43. doi:10.1186/1471-2296-13-43. PMC 3430589. PMID 22640399.
- ^ "UK 20-year vision for antimicrobial resistance". GOV.UK. Archived from the original on 28 March 2023. Retrieved 28 March 2023.
- ^ "UK 5-year action plan for antimicrobial resistance 2019 to 2024". GOV.UK. Archived from the original on 28 March 2023. Retrieved 28 March 2023.
- ^ "WHO bacterial priority pathogens list, 2024: Bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance". who.int. Archived from the original on 20 May 2024. Retrieved 20 May 2024.
- ^ "World Antibiotic Awareness Week". World Health Organization. Archived from the original on 20 November 2015. Retrieved 20 November 2015.
- ^ "World Antibiotic Awareness Week". WHO. Archived from the original on 13 November 2017. Retrieved 14 November 2017.
- ^ a b "WHO | UN Interagency Coordination Group (IACG) on Antimicrobial Resistance". WHO. Archived from the original on 20 March 2017. Retrieved 7 August 2019.
- ^ Interagency Coordination Group (IACG) (April 2019). No Time to Wait: Securing the Future from Drug-Resistant Infections (PDF) (Report). Archived (PDF) from the original on 19 April 2023. Retrieved 19 April 2023.
- ^ Reygaert WC (2018). "An overview of the antimicrobial resistance mechanisms of bacteria". AIMS Microbiology. 4 (3): 482–501. doi:10.3934/microbiol.2018.3.482. PMC 6604941. PMID 31294229.
- ^ Baylay AJ, Piddock LJ, Webber MA (14 March 2019). "Molecular Mechanisms of Antibiotic Resistance – Part I". Bacterial Resistance to Antibiotics – from Molecules to Man. Wiley. pp. 1–26. doi:10.1002/9781119593522.ch1. ISBN 978-1-119-94077-7. S2CID 202806156.
- ^ Connell SR, Tracz DM, Nierhaus KH, Taylor DE (December 2003). "Ribosomal protection proteins and their mechanism of tetracycline resistance". Antimicrobial Agents and Chemotherapy. 47 (12): 3675–81. doi:10.1128/AAC.47.12.3675-3681.2003. PMC 296194. PMID 14638464.
- ^ Henry RJ (December 1943). "The Mode of Action of Sulfonamides". Bacteriological Reviews. 7 (4): 175–262. doi:10.1128/MMBR.7.4.175-262.1943. PMC 440870. PMID 16350088.
- ^ Li XZ, Nikaido H (August 2009). "Efflux-mediated drug resistance in bacteria: an update". Drugs. 69 (12): 1555–623. doi:10.2165/11317030-000000000-00000. PMC 2847397. PMID 19678712.
- ^ Aminov RI, Mackie RI (June 2007). "Evolution and ecology of antibiotic resistance genes". FEMS Microbiology Letters. 271 (2): 147–61. doi:10.1111/j.1574-6968.2007.00757.x. PMID 17490428.
- ^ Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, et al. (July 1998). "NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli". Antimicrobial Agents and Chemotherapy. 42 (7): 1778–82. doi:10.1128/AAC.42.7.1778. PMC 105682. PMID 9661020.
- ^ Duval M, Dar D, Carvalho F, Rocha EP, Sorek R, Cossart P (December 2018). "HflXr, a homolog of a ribosome-splitting factor, mediates antibiotic resistance". Proceedings of the National Academy of Sciences of the United States of America. 115 (52): 13359–13364. Bibcode:2018PNAS..11513359D. doi:10.1073/pnas.1810555115. PMC 6310831. PMID 30545912.
- ^ "About Antibiotic Resistance". CDC. 13 March 2020. Archived from the original on 1 October 2017. Retrieved 8 September 2017.
- ^ Robicsek A, Jacoby GA, Hooper DC (October 2006). "The worldwide emergence of plasmid-mediated quinolone resistance". The Lancet. Infectious Diseases. 6 (10): 629–40. doi:10.1016/S1473-3099(06)70599-0. PMID 17008172.
- ^ Ochiai K, Yamanaka T, Kimura K, Sawada O, O (1959). "Inheritance of drug resistance (and its transfer) between Shigella strains and Between Shigella and E.coli strains". Hihon Iji Shimpor (in Japanese). 34: 1861.
- ^ Watford S, Warrington SJ (2018). "Bacterial DNA Mutations". StatPearls. StatPearls Publishing. PMID 29083710. Archived from the original on 8 March 2021. Retrieved 21 January 2019.
- ^ Levin BR, Perrot V, Walker N (March 2000). "Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria". Genetics. 154 (3): 985–97. doi:10.1093/genetics/154.3.985. PMC 1460977. PMID 10757748. Archived from the original on 18 January 2023. Retrieved 4 March 2017.
- ^ Hotchkiss RD (1951). "Transfer of penicillin resistance in pneumococci by the desoxyribonucleate derived from resistant cultures". Cold Spring Harbor Symposia on Quantitative Biology. 16: 457–61. doi:10.1101/SQB.1951.016.01.032. PMID 14942755.
- ^ Ubukata K, Konno M, Fujii R (September 1975). "Transduction of drug resistance to tetracycline, chloramphenicol, macrolides, lincomycin and clindamycin with phages induced from Streptococcus pyogenes". The Journal of Antibiotics. 28 (9): 681–8. doi:10.7164/antibiotics.28.681. PMID 1102514.
- ^ von Wintersdorff CJ, Penders J, van Niekerk JM, Mills ND, Majumder S, van Alphen LB, et al. (19 February 2016). "Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer". Frontiers in Microbiology. 7: 173. doi:10.3389/fmicb.2016.00173. PMC 4759269. PMID 26925045.
- ^ Chan CX, Beiko RG, Ragan MA (August 2011). "Lateral transfer of genes and gene fragments in Staphylococcus extends beyond mobile elements". Journal of Bacteriology. 193 (15): 3964–77. doi:10.1128/JB.01524-10. PMC 3147504. PMID 21622749.
- ^ Johansen TB, Scheffer L, Jensen VK, Bohlin J, Feruglio SL (June 2018). "Whole-genome sequencing and antimicrobial resistance in Brucella melitensis from a Norwegian perspective". Scientific Reports. 8 (1): 8538. Bibcode:2018NatSR...8.8538J. doi:10.1038/s41598-018-26906-3. PMC 5986768. PMID 29867163.
- ^ Tirumalai MR, Karouia F, Tran Q, Stepanov VG, Bruce RJ, Ott M, et al. (January 2019). "Evaluation of acquired antibiotic resistance in Escherichia coli exposed to long-term low-shear modeled microgravity and background antibiotic exposure". mBio. 10 (e02637-18). doi:10.1128/mBio.02637-18. PMC 6336426. PMID 30647159.
- ^ Tirumalai MR, Karouia F, Tran Q, Stepanov VG, Bruce RJ, Ott M, et al. (May 2017). "The adaptation of Escherichia coli cells grown in simulated microgravity for an extended period is both phenotypic and genomic". npj Microgravity. 3 (15): 15. doi:10.1038/s41526-017-0020-1. PMC 5460176. PMID 28649637.
- ^ Ghaith DM, Mohamed ZK, Farahat MG, Aboulkasem Shahin W, Mohamed HO (March 2019). "Colonization of intestinal microbiota with carbapenemase-producing Enterobacteriaceae in paediatric intensive care units in Cairo, Egypt". Arab Journal of Gastroenterology. 20 (1): 19–22. doi:10.1016/j.ajg.2019.01.002. PMID 30733176. S2CID 73444389. Archived from the original on 27 November 2022. Retrieved 30 May 2022.
- ^ Diene SM, Rolain JM (September 2014). "Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species". Clinical Microbiology and Infection. 20 (9): 831–8. doi:10.1111/1469-0691.12655. PMID 24766097.
- ^ Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. (September 2010). "Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study". The Lancet. Infectious Diseases. 10 (9): 597–602. doi:10.1016/S1473-3099(10)70143-2. PMC 2933358. PMID 20705517.
- ^ Hudson CM, Bent ZW, Meagher RJ, Williams KP (7 June 2014). "Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain". PLOS ONE. 9 (6): e99209. Bibcode:2014PLoSO...999209H. doi:10.1371/journal.pone.0099209. PMC 4048246. PMID 24905728.
- ^ Lou Z, Sun Y, Rao Z (February 2014). "Current progress in antiviral strategies". Trends in Pharmacological Sciences. 35 (2): 86–102. doi:10.1016/j.tips.2013.11.006. PMC 7112804. PMID 24439476.
- ^ Pennings PS (June 2013). "HIV Drug Resistance: Problems and Perspectives". Infectious Disease Reports. 5 (Suppl 1): e5. doi:10.4081/idr.2013.s1.e5. PMC 3892620. PMID 24470969.
- ^ Das K, Arnold E (April 2013). "HIV-1 reverse transcriptase and antiviral drug resistance. Part 1". Current Opinion in Virology. 3 (2): 111–8. doi:10.1016/j.coviro.2013.03.012. PMC 4097814. PMID 23602471.
- ^ Ton Q, Frenkel L (March 2013). "HIV drug resistance in mothers and infants following use of antiretrovirals to prevent mother-to-child transmission". Current HIV Research. 11 (2): 126–36. doi:10.2174/1570162x11311020005. PMID 23432488.
- ^ Ebrahim O, Mazanderani AH (June 2013). "Recent developments in hiv treatment and their dissemination in poor countries". Infectious Disease Reports. 5 (Suppl 1): e2. doi:10.4081/idr.2013.s1.e2. PMC 3892621. PMID 24470966.
- ^ Xie JL, Polvi EJ, Shekhar-Guturja T, Cowen LE (2014). "Elucidating drug resistance in human fungal pathogens". Future Microbiology. 9 (4): 523–42. doi:10.2217/fmb.14.18. PMID 24810351.
- ^ Srinivasan A, Lopez-Ribot JL, Ramasubramanian AK (March 2014). "Overcoming antifungal resistance". Drug Discovery Today: Technologies. 11: 65–71. doi:10.1016/j.ddtec.2014.02.005. PMC 4031462. PMID 24847655.
- ^ Costa C, Dias PJ, Sá-Correia I, Teixeira MC (2014). "MFS multidrug transporters in pathogenic fungi: do they have real clinical impact?". Frontiers in Physiology. 5: 197. doi:10.3389/fphys.2014.00197. PMC 4035561. PMID 24904431.
- ^ World Health Organization, ed. (25 October 2022). WHO fungal priority pathogens list to guide research, development and public health action. World Health Organization. ISBN 978-92-4-006024-1. Archived from the original on 26 October 2022. Retrieved 27 October 2022.
- ^ a b Andrews KT, Fisher G, Skinner-Adams TS (August 2014). "Drug repurposing and human parasitic protozoan diseases". International Journal for Parasitology: Drugs and Drug Resistance. 4 (2): 95–111. doi:10.1016/j.ijpddr.2014.02.002. PMC 4095053. PMID 25057459.
- ^ Visser BJ, van Vugt M, Grobusch MP (October 2014). "Malaria: an update on current chemotherapy". Expert Opinion on Pharmacotherapy. 15 (15): 2219–54. doi:10.1517/14656566.2014.944499. PMID 25110058. S2CID 34991324.
- ^ Chia WN, Goh YS, Rénia L (2014). "Novel approaches to identify protective malaria vaccine candidates". Frontiers in Microbiology. 5: 586. doi:10.3389/fmicb.2014.00586. PMC 4233905. PMID 25452745.
- ^ Franco JR, Simarro PP, Diarra A, Jannin JG (2014). "Epidemiology of human African trypanosomiasis". Clinical Epidemiology. 6: 257–75. doi:10.2147/CLEP.S39728. PMC 4130665. PMID 25125985.
- ^ Herrera L (2014). "Trypanosoma cruzi, the Causal Agent of Chagas Disease: Boundaries between Wild and Domestic Cycles in Venezuela". Frontiers in Public Health. 2: 259. doi:10.3389/fpubh.2014.00259. PMC 4246568. PMID 25506587.
- ^ Mansueto P, Seidita A, Vitale G, Cascio A (2014). "Leishmaniasis in travelers: a literature review" (PDF). Travel Medicine and Infectious Disease. 12 (6 Pt A): 563–81. doi:10.1016/j.tmaid.2014.09.007. hdl:10447/101959. PMID 25287721. Archived (PDF) from the original on 12 October 2022. Retrieved 23 October 2017.
- ^ a b c Munk P, Brinch C, Møller FD, Petersen TN, Hendriksen RS, Seyfarth AM, et al. (December 2022). "Genomic analysis of sewage from 101 countries reveals global landscape of antimicrobial resistance". Nature Communications. 13 (1): 7251. Bibcode:2022NatCo..13.7251M. doi:10.1038/s41467-022-34312-7. PMC 9715550. PMID 36456547.
- ^ "Antibiotika-Resistenzen verbreiten sich offenbar anders als gedacht". Deutschlandfunk Nova (in German). Archived from the original on 17 January 2023. Retrieved 17 January 2023.
- ^ "Superbugs on the rise: WHO report signals increase in antibiotic resistance". World Health Organization via medicalxpress.com. Archived from the original on 2 February 2023. Retrieved 18 January 2023.
- ^ "Global antimicrobial resistance and use surveillance system (GLASS) report: 2022". who.int. Archived from the original on 21 January 2023. Retrieved 18 January 2023.
- ^ "Patients contracted 165 antibiotic resistant infections each day in 2018, says PHE". Pharmaceutical Journal. 31 October 2019. Archived from the original on 26 July 2020. Retrieved 11 December 2019.
- ^ "National Infection & Death Estimates for AR". Centers for Disease Control and Prevention. 6 July 2022. Archived from the original on 3 August 2023. Retrieved 3 August 2023.
- ^ "COVID-19 & Antibiotic Resistance". Centers for Disease Control and Prevention. 11 August 2022. Archived from the original on 21 February 2022. Retrieved 3 August 2023.
- ^ "2022 SPECIAL REPORT: COVID-19 U.S. IMPACT ON ANTIMICROBIAL RESISTANCE" (PDF). Centers for Disease Control and Prevention. 2022. p. 7. Archived (PDF) from the original on 26 August 2023. Retrieved 3 August 2023.
- ^ Aminov RI (2010). "A brief history of the antibiotic era: lessons learned and challenges for the future". Frontiers in Microbiology. 1: 134. doi:10.3389/fmicb.2010.00134. PMC 3109405. PMID 21687759.
- ^ Carvalho G, Forestier C, Mathias JD (December 2019). "Antibiotic resilience: a necessary concept to complement antibiotic resistance?". Proceedings. Biological Sciences. 286 (1916): 20192408. doi:10.1098/rspb.2019.2408. PMC 6939251. PMID 31795866.
- ^ a b Antimicrobial resistance : global report on surveillance. Geneva, Switzerland: World Health Organization. 2014. ISBN 978-92-4-156474-8. OCLC 880847527.
- ^ Amábile-Cuevas CF, ed. (2007). Antimicrobial resistance in bacteria. Horizon Scientific Press.
- ^ Fleming A (11 December 1945). "Penicillin" (PDF). Nobel Lecture. Archived (PDF) from the original on 31 March 2018. Retrieved 9 August 2020.
- ^ "WHO | Global action plan on antimicrobial resistance". WHO. Archived from the original on 18 April 2018. Retrieved 23 April 2018.
- ^ a b Pollack A (20 January 2016). "To Fight 'Superbugs,' Drug Makers Call for Incentives to Develop Antibiotics". New York Times. Davos 2016 Special Report. Davos, Switzerland. Archived from the original on 24 April 2018. Retrieved 24 January 2016.
- ^ Leonard C, Crabb N, Glover D, Cooper S, Bouvy J, Wobbe M, et al. (May 2023). "Can the UK 'Netflix' Payment Model Boost the Antibacterial Pipeline?". Applied Health Economics and Health Policy. 21 (3): 365–372. doi:10.1007/s40258-022-00786-1. PMC 9842493. PMID 36646872.
- ^ a b Behdinan A, Hoffman SJ, Pearcey M (2015). "Some Global Policies for Antibiotic Resistance Depend on Legally Binding and Enforceable Commitments". The Journal of Law, Medicine & Ethics. 43 (2 Suppl 3): 68–73. doi:10.1111/jlme.12277. PMID 26243246. S2CID 7415203.
- ^ Hoffman SJ, Outterson K (2015). "What Will It Take to Address the Global Threat of Antibiotic Resistance?". The Journal of Law, Medicine & Ethics. 43 (2): 363–8. doi:10.1111/jlme.12253. PMID 26242959. S2CID 41987305. Archived from the original on 12 October 2022. Retrieved 11 November 2019.
- ^ a b c Hoffman SJ, Outterson K, Røttingen JA, Cars O, Clift C, Rizvi Z, et al. (February 2015). "An international legal framework to address antimicrobial resistance". Bulletin of the World Health Organization. 93 (2): 66. doi:10.2471/BLT.15.152710 (inactive 9 November 2024). PMC 4339972. PMID 25883395.
{{cite journal}}: CS1 maint: DOI inactive as of November 2024 (link) - ^ Rizvi Z, Hoffman SJ (2015). "Effective Global Action on Antibiotic Resistance Requires Careful Consideration of Convening Forums". The Journal of Law, Medicine & Ethics. 43 (2 Suppl 3): 74–8. doi:10.1111/jlme.12278. PMID 26243247. S2CID 24223063.
- ^ Andresen S, Hoffman SJ (2015). "Much Can Be Learned about Addressing Antibiotic Resistance from Multilateral Environmental Agreements". Journal of Law, Medicine & Ethics. 43 (2): 46–52.
- ^ President's 2016 Budget Proposes Historic Investment to Combat Antibiotic-Resistant Bacteria to Protect Public Health Archived 22 January 2017 at the Wayback Machine The White House, Office of the Press Secretary, 27 January 2015
- ^ a b "FACT SHEET: Obama Administration Releases National Action Plan to Combat Antibiotic-Resistant Bacteria". whitehouse.gov. 27 March 2015. Archived from the original on 21 January 2017. Retrieved 30 October 2015 – via National Archives.
- ^ a b "Antibiotic Resistance Threats in the United States, 2023". Centers for Disease Control and Prevention. Retrieved 20 October 2023.
- ^ Petrakis V, Panopoulou M, Rafailidis P, Lemonakis N, Lazaridis G, Terzi I, et al. (2023). "The Impact of the COVID-19 Pandemic on Antimicrobial Resistance and Management of Bloodstream Infections". Pathogens. 21 (6): e22 – e25. doi:10.3390/pathogens12060780. PMC 10302285. PMID 37375470.
- ^ "Get Ahead of Sepsis". Centers for Disease Control and Prevention. Retrieved 20 October 2023.
- ^ "National Action Plan for Combating Antibiotic-Resistant Bacteria (CARB) 2023-2028". The White House. Retrieved 20 October 2023.
- ^ "PASTEUR Act of 2023". United States Congress. 28 October 2021. Retrieved 20 October 2023.
- ^ Knight GM, Davies NG, Colijn C, Coll F, Donker T, Gifford DR, et al. (November 2019). "Mathematical modelling for antibiotic resistance control policy: do we know enough?". BMC Infectious Diseases. 19 (1): 1011. doi:10.1186/s12879-019-4630-y. PMC 6884858. PMID 31783803.
- ^ Hopkins H, Bruxvoort KJ, Cairns ME, Chandler CI, Leurent B, Ansah EK, et al. (March 2017). "Impact of introduction of rapid diagnostic tests for malaria on antibiotic prescribing: analysis of observational and randomised studies in public and private healthcare settings". BMJ. 356: j1054. doi:10.1136/bmj.j1054. PMC 5370398. PMID 28356302.
- ^ "Diagnostics Are Helping Counter Antimicrobial Resistance, But More Work Is Needed". MDDI Online. 20 November 2018. Archived from the original on 2 December 2018. Retrieved 2 December 2018.
- ^ a b van Belkum A, Bachmann TT, Lüdke G, Lisby JG, Kahlmeter G, Mohess A, et al. (January 2019). "Developmental roadmap for antimicrobial susceptibility testing systems". Nature Reviews. Microbiology. 17 (1): 51–62. doi:10.1038/s41579-018-0098-9. hdl:2445/132505. PMC 7138758. PMID 30333569.
- ^ "Progress on antibiotic resistance". Nature. 562 (7727): 307. October 2018. Bibcode:2018Natur.562Q.307.. doi:10.1038/d41586-018-07031-7. PMID 30333595.
- ^ Kim JI, Maguire F, Tsang KK, Gouliouris T, Peacock SJ, McAllister TA, et al. (September 2022). "Machine Learning for Antimicrobial Resistance Prediction: Current Practice, Limitations, and Clinical Perspective". Clinical Microbiology Reviews. 35 (3): e0017921. doi:10.1128/cmr.00179-21. PMC 9491192. PMID 35612324.
- ^ Banerjee R, Patel R (February 2023). "Molecular diagnostics for genotypic detection of antibiotic resistance: current landscape and future directions". JAC-antimicrobial Resistance. 5 (1): dlad018. doi:10.1093/jacamr/dlad018. PMC 9937039. PMID 36816746.
- ^ Baltekin Ö, Boucharin A, Tano E, Andersson DI, Elf J (August 2017). "Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging". Proceedings of the National Academy of Sciences of the United States of America. 114 (34): 9170–9175. Bibcode:2017PNAS..114.9170B. doi:10.1073/pnas.1708558114. PMC 5576829. PMID 28790187.
- ^ Luro S, Potvin-Trottier L, Okumus B, Paulsson J (January 2020). "Isolating live cells after high-throughput, long-term, time-lapse microscopy". Nature Methods. 17 (1): 93–100. doi:10.1038/s41592-019-0620-7. PMC 7525750. PMID 31768062.
- ^ Schuetz P, Wirz Y, Sager R, Christ-Crain M, Stolz D, Tamm M, et al. (Cochrane Acute Respiratory Infections Group) (October 2017). "Procalcitonin to initiate or discontinue antibiotics in acute respiratory tract infections". The Cochrane Database of Systematic Reviews. 10 (10): CD007498. doi:10.1002/14651858.CD007498.pub3. PMC 6485408. PMID 29025194.
- ^ Smedemark SA, Aabenhus R, Llor C, Fournaise A, Olsen O, Jørgensen KJ, et al. (Cochrane Acute Respiratory Infections Group) (October 2022). "Biomarkers as point-of-care tests to guide prescription of antibiotics in people with acute respiratory infections in primary care". The Cochrane Database of Systematic Reviews. 2022 (10): CD010130. doi:10.1002/14651858.CD010130.pub3. PMC 9575154. PMID 36250577.
- ^ Doan Q, Enarson P, Kissoon N, Klassen TP, Johnson DW (September 2014). "Rapid viral diagnosis for acute febrile respiratory illness in children in the Emergency Department". The Cochrane Database of Systematic Reviews. 2014 (9): CD006452. doi:10.1002/14651858.CD006452.pub4. PMC 6718218. PMID 25222468.
- ^ Mishra RP, Oviedo-Orta E, Prachi P, Rappuoli R, Bagnoli F (October 2012). "Vaccines and antibiotic resistance". Current Opinion in Microbiology. 15 (5): 596–602. doi:10.1016/j.mib.2012.08.002. PMID 22981392.
- ^ Kennedy DA, Read AF (March 2017). "Why does drug resistance readily evolve but vaccine resistance does not?". Proceedings. Biological Sciences. 284 (1851): 20162562. doi:10.1098/rspb.2016.2562. PMC 5378080. PMID 28356449.
- ^ Kennedy DA, Read AF (December 2018). "Why the evolution of vaccine resistance is less of a concern than the evolution of drug resistance". Proceedings of the National Academy of Sciences of the United States of America. 115 (51): 12878–12886. Bibcode:2018PNAS..11512878K. doi:10.1073/pnas.1717159115. PMC 6304978. PMID 30559199.
- ^ "Immunity, Infectious Diseases, and Pandemics—What You Can Do". HomesteadSchools.com. Archived from the original on 3 December 2013. Retrieved 12 June 2013.
- ^ Shinefield H, Black S, Fattom A, Horwith G, Rasgon S, Ordonez J, et al. (February 2002). "Use of a Staphylococcus aureus conjugate vaccine in patients receiving hemodialysis". The New England Journal of Medicine. 346 (7): 491–6. doi:10.1056/NEJMoa011297. PMID 11844850.
- ^ a b Fowler VG, Proctor RA (May 2014). "Where does a Staphylococcus aureus vaccine stand?". Clinical Microbiology and Infection. 20 (5): 66–75. doi:10.1111/1469-0691.12570. PMC 4067250. PMID 24476315.
- ^ McNeely TB, Shah NA, Fridman A, Joshi A, Hartzel JS, Keshari RS, et al. (2014). "Mortality among recipients of the Merck V710 Staphylococcus aureus vaccine after postoperative S. aureus infections: an analysis of possible contributing host factors". Human Vaccines & Immunotherapeutics. 10 (12): 3513–6. doi:10.4161/hv.34407. PMC 4514053. PMID 25483690.
- ^ Lu RM, Hwang YC, Liu IJ, Lee CC, Tsai HZ, Li HJ, et al. (January 2020). "Development of therapeutic antibodies for the treatment of diseases". Journal of Biomedical Science. 27 (1): 1. doi:10.1186/s12929-019-0592-z. PMC 6939334. PMID 31894001.
- ^ a b c Singh G, Rana A (May 2024). "Decoding antimicrobial resistance: unraveling molecular mechanisms and targeted strategies". Archives of Microbiology. 206 (6): 280. Bibcode:2024ArMic.206..280S. doi:10.1007/s00203-024-03998-2. PMID 38805035.
- ^ Kim S, Lieberman TD, Kishony R (October 2014). "Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance". Proceedings of the National Academy of Sciences of the United States of America. 111 (40): 14494–9. Bibcode:2014PNAS..11114494K. doi:10.1073/pnas.1409800111. PMC 4210010. PMID 25246554.
- ^ Pál C, Papp B, Lázár V (July 2015). "Collateral sensitivity of antibiotic-resistant microbes". Trends in Microbiology. 23 (7): 401–7. doi:10.1016/j.tim.2015.02.009. PMC 5958998. PMID 25818802.
- ^ Imamovic L, Ellabaan MM, Dantas Machado AM, Citterio L, Wulff T, Molin S, et al. (January 2018). "Drug-Driven Phenotypic Convergence Supports Rational Treatment Strategies of Chronic Infections". Cell. 172 (1–2): 121–134.e14. doi:10.1016/j.cell.2017.12.012. PMC 5766827. PMID 29307490.
- ^ Beckley AM, Wright ES (October 2021). "Identification of antibiotic pairs that evade concurrent resistance via a retrospective analysis of antimicrobial susceptibility test results". The Lancet. Microbe. 2 (10): e545 – e554. doi:10.1016/S2666-5247(21)00118-X. PMC 8496867. PMID 34632433.
- ^ Ma Y, Chua SL (15 November 2021). "No collateral antibiotic sensitivity by alternating antibiotic pairs". The Lancet Microbe. 3 (1): e7. doi:10.1016/S2666-5247(21)00270-6. ISSN 2666-5247. PMID 35544116. S2CID 244147577.
- ^ Liu J, Bedell TA, West JG, Sorensen EJ (June 2016). "Design and Synthesis of Molecular Scaffolds with Anti-infective Activity". Tetrahedron. 72 (25): 3579–3592. doi:10.1016/j.tet.2016.01.044. PMC 4894353. PMID 27284210.
- ^ "Annual Report of the Chief Medical Officer - Infections and the rise of antimicrobial resistance" (PDF). UK NHS. 2011. Archived from the original (PDF) on 30 October 2013.
- ^ "Obama Administration Seeks To Ease Approvals For Antibiotics". NPR.org. NPR. 4 June 2013. Archived from the original on 13 March 2015. Retrieved 7 August 2016.
- ^ Prayle A, Watson A, Fortnum H, Smyth A (July 2010). "Side effects of aminoglycosides on the kidney, ear and balance in cystic fibrosis". Thorax. 65 (7): 654–658. doi:10.1136/thx.2009.131532. PMC 2921289. PMID 20627927.
- ^ Alene KA, Wangdi K, Colquhoun S, Chani K, Islam T, Rahevar K, et al. (September 2021). "Tuberculosis related disability: a systematic review and meta-analysis". BMC Medicine. 19 (1): 203. doi:10.1186/s12916-021-02063-9. PMC 8426113. PMID 34496845.
- ^ a b Walsh F (11 March 2013). "BBC News – Antibiotics resistance 'as big a risk as terrorism' – medical chief". BBC News. Bbc.co.uk. Archived from the original on 8 August 2018. Retrieved 12 March 2013.
- ^ a b Berida T, Adekunle Y, Dada-Adegbola H, Kdimy A, Roy S, Sarker S (May 2024). "Plant antibacterials: The challenges and opportunities". Heliyon. 10 (10): e31145. Bibcode:2024Heliy..1031145B. doi:10.1016/j.heliyon.2024.e31145. PMC 11128932. PMID 38803958.
- ^ Khor M (18 May 2014). "Why Are Antibiotics Becoming Useless All Over the World?". The Real News. Archived from the original on 18 May 2014. Retrieved 18 May 2014.
- ^ Nordrum A (2015). "Antibiotic Resistance: Why Aren't Drug Companies Developing New Medicines to Stop Superbugs?". International Business Times.
- ^ Gever J (4 February 2011). "Pfizer Moves May Dim Prospect for New Antibiotics". MedPage Today. Archived from the original on 14 December 2013. Retrieved 12 March 2013.
- ^ Schmitt S, Montalbán-López M, Peterhoff D, Deng J, Wagner R, Held M, et al. (May 2019). "Analysis of modular bioengineered antimicrobial lanthipeptides at nanoliter scale" (PDF). Nature Chemical Biology. 15 (5): 437–443. doi:10.1038/s41589-019-0250-5. PMID 30936500. S2CID 91188986. Archived (PDF) from the original on 18 April 2023. Retrieved 12 April 2023.
- ^ "Overview". Carb-X. Archived from the original on 28 March 2023. Retrieved 28 March 2023.
- ^ "Novel Gram Negative Antibiotic Now". CORDIS. doi:10.3030/853979. Archived from the original on 28 March 2023. Retrieved 28 March 2023.
- ^ "About". REPAIR Impact Fund. Archived from the original on 23 January 2019. Retrieved 28 March 2023.
- ^ "ADVANcing Clinical Evidence in Infectious Diseases (ADVANCE-ID)". sph.nus.edu.sg. Archived from the original on 28 March 2023. Retrieved 28 March 2023.
- ^ "About GARDP". GARDP. 27 September 2022. Archived from the original on 28 March 2023. Retrieved 28 March 2023.
- ^ Hoenigl M, Sprute R, Egger M, Arastehfar A, Cornely OA, Krause R, et al. (October 2021). "The Antifungal Pipeline: Fosmanogepix, Ibrexafungerp, Olorofim, Opelconazole, and Rezafungin". Drugs. 81 (15): 1703–1729. doi:10.1007/s40265-021-01611-0. PMC 8501344. PMID 34626339.
- ^ "Antibiotic resistance outwitted by supercomputers". University of Portsmouth. Archived from the original on 13 December 2021. Retrieved 13 December 2021.
- ^ König G, Sokkar P, Pryk N, Heinrich S, Möller D, Cimicata G, et al. (November 2021). "Rational prioritization strategy allows the design of macrolide derivatives that overcome antibiotic resistance". Proceedings of the National Academy of Sciences of the United States of America. 118 (46): e2113632118. Bibcode:2021PNAS..11813632K. doi:10.1073/pnas.2113632118. PMC 8609559. PMID 34750269.
- ^ Leppäranta O, Vaahtio M, Peltola T, Zhang D, Hupa L, Hupa M, et al. (February 2008). "Antibacterial effect of bioactive glasses on clinically important anaerobic bacteria in vitro". Journal of Materials Science. Materials in Medicine. 19 (2): 547–51. doi:10.1007/s10856-007-3018-5. PMID 17619981. S2CID 21444777.
- ^ Munukka E, Leppäranta O, Korkeamäki M, Vaahtio M, Peltola T, Zhang D, et al. (January 2008). "Bactericidal effects of bioactive glasses on clinically important aerobic bacteria". Journal of Materials Science. Materials in Medicine. 19 (1): 27–32. doi:10.1007/s10856-007-3143-1. PMID 17569007. S2CID 39643380.
- ^ Drago L, De Vecchi E, Bortolin M, Toscano M, Mattina R, Romanò CL (August 2015). "Antimicrobial activity and resistance selection of different bioglass S53P4 formulations against multidrug resistant strains". Future Microbiology. 10 (8): 1293–9. doi:10.2217/FMB.15.57. PMID 26228640.
- ^ Ermini ML, Voliani V (April 2021). "Antimicrobial Nano-Agents: The Copper Age". ACS Nano. 15 (4): 6008–6029. doi:10.1021/acsnano.0c10756. PMC 8155324. PMID 33792292.
- ^ Connelly E (18 April 2017). "Medieval medical books could hold the recipe for new antibiotics". The Conversation. Archived from the original on 25 December 2022. Retrieved 20 December 2019.
- ^ "AncientBiotics – a medieval remedy for modern day superbugs?" (Press release). University of Nottingham. 30 March 2015. Archived from the original on 21 October 2022. Retrieved 13 August 2019.
- ^ a b Kim DW, Cha CJ (March 2021). "Antibiotic resistome from the One-Health perspective: understanding and controlling antimicrobial resistance transmission". Experimental & Molecular Medicine. 53 (3): 301–309. doi:10.1038/s12276-021-00569-z. PMC 8080597. PMID 33642573.
- ^ Alcock BP, Huynh W, Chalil R, Smith KW, Raphenya AR, Wlodarski MA, et al. (January 2023). "CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database". Nucleic Acids Research. 51 (D1): D690 – D699. doi:10.1093/nar/gkac920. PMC 9825576. PMID 36263822.
- ^ "The Comprehensive Antibiotic Resistance Database". card.mcmaster.ca. Archived from the original on 28 March 2023. Retrieved 28 March 2023.
- ^ Florensa AF, Kaas RS, Clausen PT, Aytan-Aktug D, Aarestrup FM (January 2022). "ResFinder – an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes". Microbial Genomics. 8 (1). doi:10.1099/mgen.0.000748. PMC 8914360. PMID 35072601.
- ^ "ResFinder 4.1". cge.food.dtu.dk. Archived from the original on 28 March 2023. Retrieved 28 March 2023.
- ^ "Silent Killers: Fantastic Phages?". CBS News. 19 September 2002. Archived from the original on 10 February 2013. Retrieved 14 November 2017.
- ^ McAuliffe O, Ross RP, Fitzgerald GF (2007). "The New Phage Biology: From Genomics to Applications" (introduction)". In McGrath S, van Sinderen D (eds.). Bacteriophage: Genetics and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-14-1. Archived from the original on 26 November 2020. Retrieved 11 August 2021.
- ^ Lin DM, Koskella B, Lin HC (August 2017). "Phage therapy: An alternative to antibiotics in the age of multi-drug resistance". World Journal of Gastrointestinal Pharmacology and Therapeutics. 8 (3): 162–173. doi:10.4292/wjgpt.v8.i3.162. PMC 5547374. PMID 28828194.
- ^ a b Salmond GP, Fineran PC (December 2015). "A century of the phage: past, present and future". Nature Reviews. Microbiology. 13 (12): 777–86. doi:10.1038/nrmicro3564. PMID 26548913. S2CID 8635034.
- ^ Letarov AV, Golomidova AK, Tarasyan KK (April 2010). "Ecological basis for rational phage therapy". Acta Naturae. 2 (1): 60–72. doi:10.32607/20758251-2010-2-1-60-71. PMC 3347537. PMID 22649629.
- ^ Parfitt T (June 2005). "Georgia: an unlikely stronghold for bacteriophage therapy". Lancet. 365 (9478): 2166–7. doi:10.1016/S0140-6736(05)66759-1. PMID 15986542. S2CID 28089251.
- ^ Golkar Z, Bagasra O, Pace DG (February 2014). "Bacteriophage therapy: a potential solution for the antibiotic resistance crisis". Journal of Infection in Developing Countries. 8 (2): 129–36. doi:10.3855/jidc.3573. PMID 24518621.
- ^ McCallin S, Alam Sarker S, Barretto C, Sultana S, Berger B, Huq S, et al. (September 2013). "Safety analysis of a Russian phage cocktail: from metagenomic analysis to oral application in healthy human subjects". Virology. 443 (2): 187–96. doi:10.1016/j.virol.2013.05.022. PMID 23755967.
- ^ Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM (March 2011). "Phage treatment of human infections". Bacteriophage. 1 (2): 66–85. doi:10.4161/bact.1.2.15845. PMC 3278644. PMID 22334863.
- ^ Onsea J, Soentjens P, Djebara S, Merabishvili M, Depypere M, Spriet I, et al. (September 2019). "Bacteriophage Application for Difficult-to-treat Musculoskeletal Infections: Development of a Standardized Multidisciplinary Treatment Protocol". Viruses. 11 (10): 891. doi:10.3390/v11100891. PMC 6832313. PMID 31548497.
Books
[edit]- Caldwell R, Lindberg D, eds. (2011). "Understanding Evolution" [Mutations are random]. University of California Museum of Paleontology. Archived from the original on 8 February 2012. Retrieved 15 August 2011.
- Reynolds LA, Tansey EM, eds. (2008). Superbugs and superdrugs : a history of MRSA : the transcript of a Witness Seminar held by the Wellcome Trust Centre for the History of Medicine at UCL, London, on 11 July 2006. London: Wellcome Trust Centre for the History of Medicine at UCL. ISBN 978-0-85484-114-1.
- Stemming the Superbug Tide: Just A Few Dollars More. OECD Health Policy Studies. Paris: OECD Publishing. 2018. doi:10.1787/9789264307599-en. ISBN 978-92-64-30758-2. S2CID 239804815.
Journals
[edit]- Arias CA, Murray BE (January 2009). "Antibiotic-resistant bugs in the 21st century – a clinical super-challenge". The New England Journal of Medicine. 360 (5): 439–443. doi:10.1056/NEJMp0804651. PMID 19179312. S2CID 205104375.
- "Special Issue: Ethics and Antimicrobial Resistance". Bioethics. 365 (33). 2019. Archived from the original on 9 March 2022. Retrieved 22 January 2020.
- Goossens H, Ferech M, Vander Stichele R, Elseviers M (2005). "Outpatient antibiotic use in Europe and association with resistance: a cross-national database study". Lancet. Group Esac Project. 365 (9459). Esac Project: 579–587. doi:10.1016/S0140-6736(05)17907-0. PMID 15708101. S2CID 23782228.
- Hawkey PM, Jones AM (September 2009). "The changing epidemiology of resistance" (PDF). The Journal of Antimicrobial Chemotherapy. 64 (Suppl 1): i3–10. doi:10.1093/jac/dkp256. PMID 19675017. Archived from the original on 17 February 2024. Retrieved 20 April 2018.
- Soulsby EJ (November 2005). "Resistance to antimicrobials in humans and animals". BMJ. 331 (7527): 1219–1220. doi:10.1136/bmj.331.7527.1219. PMC 1289307. PMID 16308360.
- "Alternatives to Antibiotics Reduce Animal Disease". Commonwealth Scientific and Industrial Research Organization. 9 January 2006. Archived from the original on 5 June 2011. Retrieved 26 April 2009.
- Cooke P, Rees-Roberts D (2017). CATCH. Archived from the original on 9 March 2022. Retrieved 23 February 2017. 16-minute film about a post-antibiotic world. Review: Sansom C (March 2017). "Media Watch: An intimate family story in a world without antibiotics". Lancet Infect Dis. 17 (3): 274. doi:10.1016/S1473-3099(17)30067-1.
Further reading
[edit]- Bancroft, EA (October 2007). "Antimicrobial resistance: it's not just for hospitals". JAMA. 298 (15): 1803–1804. doi:10.1001/jama.298.15.1803. PMC 2536104. PMID 17940239.
- Larson, E (2007). "Community factors in the development of antibiotic resistance". Annual Review of Public Health. 28 (1): 435–447. doi:10.1146/annurev.publhealth.28.021406.144020. PMID 17094768.
External links
[edit] Quotations related to Antimicrobial resistance at Wikiquote
Quotations related to Antimicrobial resistance at Wikiquote- WHO fact sheet on antimicrobial resistance
- Animation of Antibiotic Resistance Archived 28 September 2022 at the Wayback Machine
- Bracing for Superbugs: Strengthening environmental action in the One Health response to antimicrobial resistance UNEP, 2023.
- CDC Guideline "Management of Multidrug-Resistant Organisms in Healthcare Settings, 2006"
