Dockerin: Difference between revisions
mNo edit summary |
Citation bot (talk | contribs) Add: s2cid, doi-access. | Use this bot. Report bugs. | Suggested by Headbomb | Linked from Wikipedia:WikiProject_Academic_Journals/Journals_cited_by_Wikipedia/Sandbox | #UCB_webform_linked 710/958 |
||
| (16 intermediate revisions by 13 users not shown) | |||
| Line 13: | Line 13: | ||
| OPM family = |
| OPM family = |
||
| OPM protein = |
| OPM protein = |
||
| CDD = cd14253 |
|||
| PDB = |
| PDB = |
||
}} |
}} |
||
| ⚫ | '''Dockerin''' is a [[protein domain]] found in the [[cellulosome]] cellular structure of [[anaerobic bacteria]]. It is found on many [[endoglucanase]] enzymes. The dockerin's binding partner is the [[cohesin domain|cohesin]] domain, located on the [[scaffoldin]] [[protein]]. This interaction between the dockerin domains of the [[enzyme]] constituents of the cellulosome and the cohesin domains of the scaffoldin protein is essential to the construction of the cellulosome complex.<ref name="pmid11222592">{{cite journal| vauthors=Ding SY, Rincon MT, Lamed R, Martin JC, McCrae SI, Aurilia V| title=Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. | journal=J Bacteriol | year= 2001 | volume= 183 | issue= 6 | pages= 1945–53 | pmid=11222592 | doi=10.1128/JB.183.6.1945-1953.2001 | pmc=95089 |display-authors=etal}}</ref> The Dockerin domain has two in-tandem repeats of a non-[[EF hand]] calcium binding motif. Each motif is characterized by a [[loop-helix structure]].<ref>{{SCOP|63447}}</ref> The three-dimensional structure of dockerin has been determined in solution,<ref name="pmid11273698">{{PDB|1DAQ}}; {{cite journal | vauthors = Lytle BL, Volkman BF, Westler WM, Heckman MP, Wu JH | title = Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain | journal = J. Mol. Biol. | volume = 307 | issue = 3 | pages = 745–53 |date=March 2001 | pmid = 11273698 | doi = 10.1006/jmbi.2001.4522 }}</ref> as well as in complex with Cohesin.<ref name="pmid14623971">{{PDB|1OHZ}}; {{cite journal|last1=Carvalho|first1=A. L.|last2=Dias|first2=F. M. V.|last3=Prates|first3=J. A. M.|last4=Nagy|first4=T.|last5=Gilbert|first5=H. J.|author-link5=Harry J. Gilbert|last6=Davies|first6=G. J.|author-link6=Gideon Davies|last7=Ferreira|first7=L. M. A.|last8=Romao|first8=M. J.|last9=Fontes|first9=C. M. G. A.|title=Cellulosome assembly revealed by the crystal structure of the cohesin-dockerin complex|journal=Proceedings of the National Academy of Sciences|volume=100|issue=24|year=2003|pages=13809–13814|doi=10.1073/pnas.1936124100|pmid = 14623971 | pmc = 283503|doi-access=free}}</ref> |
||
{{Refimprove|date=June 2008}} |
|||
| ⚫ | '''Dockerin''' is a [[protein domain]] found in the [[ |
||
There are three types of Dockerin domains: I, II and III which bind to Cohesin Type I, Cohesin Type II and Cohesin Type III respectively. A type I dockerin domain is 65-70 [[residue (chemistry)|residue]]s long.<ref>[[InterPro]]: {{InterPro|IPR016134}}</ref> The binding specificity of Type I interaction was well studied by structural and [[mutagenesis]] studies. Type II interaction is less well characterized.<ref name="pmid15697243">{{cite journal | |
There are three types of Dockerin domains: I, II and III which bind to Cohesin Type I, Cohesin Type II and Cohesin Type III respectively. A type I dockerin domain is 65-70 [[residue (chemistry)|residue]]s long.<ref>[[InterPro]]: {{InterPro|IPR016134}}</ref> The binding specificity of Type I interaction was well studied by structural and [[mutagenesis]] studies. Type II interaction is less well characterized.<ref name="pmid15697243">{{cite journal | vauthors = Adams JJ, Webb BA, Spencer HL, Smith SP | title = Structural characterization of type II dockerin module from the cellulosome of Clostridium thermocellum: calcium-induced effects on conformation and target recognition | journal = Biochemistry | volume = 44 | issue = 6 | pages = 2173–82 |date=February 2005 | pmid = 15697243 | doi = 10.1021/bi048039u }}</ref> |
||
==See also== |
==See also== |
||
| Line 31: | Line 31: | ||
Protein Structure: |
Protein Structure: |
||
* {{cite journal | |
* {{cite journal |vauthors=Lytle BL, Volkman BF, Westler WM, Heckman MP, Wu JH|title=Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain|journal=J Mol Biol|volume=307|pages=745–753|year=2001 |pmid=11273698 |doi=10.1006/jmbi.2001.4522 |issue=3 }} |
||
* {{cite journal | |
* {{cite journal |vauthors=Bayer EA, Shimon LJ, Shoham Y, Lamed R|title=Cellulosomes-structure and ultrastructure|journal=J Struct Biol|volume=124|issue=2–3|pages=221–234|year=1998|pmid=10049808 |doi=10.1006/jsbi.1998.4065}} |
||
Specificity Characterization: |
Specificity Characterization: |
||
* {{cite journal | |
* {{cite journal |vauthors=Haimovitz R, Barak Y, Morag E, Voronov-Goldman M, Shoham Y, Lamed R, Bayer EA|title=Cohesin-dockerin microarray: Diverse specificities between two complementary families of interacting protein modules|journal=Proteomics|volume=8|issue=5|pages=968–979|year=2008 |pmid=18219699 |doi=10.1002/pmic.200700486|s2cid=206360888}} |
||
* {{cite journal | |
* {{cite journal |vauthors=Adams JJ, Webb BA, Spencer HL, Smith SP|title=Structural characterization of type II dockerin module from the cellulosome of Clostridium thermocellum: calcium-induced effects on conformation and target recognition|journal=Biochemistry|volume=44|issue=6|pages=2173–2182 |year=2005 |pmid=15697243 |doi=10.1021/bi048039u}} |
||
* {{cite journal | |
* {{cite journal |vauthors=Jindou S, Soda A, Karita S, Kajino T, Béguin P, Wu JH, Inagaki M, Kimura T, Sakka K, Ohmiya K|title=Cohesin-dockerin interactions within and between Clostridium josui and Clostridium thermocellum: binding selectivity between cognate dockerin and cohesin domains and species specificity|journal=J Biol Chem|volume=279|issue=11|pages=9867–9874|year=2004 |pmid=14688277 |doi=10.1074/jbc.M308673200|doi-access=free}} |
||
[[Category: |
[[Category:Protein structural motifs]] |
||
[[Category:Protein domains]] |
[[Category:Protein domains]] |
||
Latest revision as of 09:34, 27 August 2021
| Dockerin domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
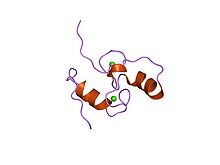 Structure of the Dockerin type I domain from C. thermocellum cellulosome. | |||||||||
| Identifiers | |||||||||
| Symbol | Dockerin_1 | ||||||||
| Pfam | PF00404 | ||||||||
| InterPro | IPR018242 | ||||||||
| PROSITE | PDOC00416 | ||||||||
| SCOP2 | 1daq / SCOPe / SUPFAM | ||||||||
| CDD | cd14253 | ||||||||
| |||||||||
Dockerin is a protein domain found in the cellulosome cellular structure of anaerobic bacteria. It is found on many endoglucanase enzymes. The dockerin's binding partner is the cohesin domain, located on the scaffoldin protein. This interaction between the dockerin domains of the enzyme constituents of the cellulosome and the cohesin domains of the scaffoldin protein is essential to the construction of the cellulosome complex.[1] The Dockerin domain has two in-tandem repeats of a non-EF hand calcium binding motif. Each motif is characterized by a loop-helix structure.[2] The three-dimensional structure of dockerin has been determined in solution,[3] as well as in complex with Cohesin.[4]
There are three types of Dockerin domains: I, II and III which bind to Cohesin Type I, Cohesin Type II and Cohesin Type III respectively. A type I dockerin domain is 65-70 residues long.[5] The binding specificity of Type I interaction was well studied by structural and mutagenesis studies. Type II interaction is less well characterized.[6]
See also
[edit]References
[edit]- ^ Ding SY, Rincon MT, Lamed R, Martin JC, McCrae SI, Aurilia V, et al. (2001). "Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens". J Bacteriol. 183 (6): 1945–53. doi:10.1128/JB.183.6.1945-1953.2001. PMC 95089. PMID 11222592.
- ^ SCOP 63447
- ^ PDB: 1DAQ; Lytle BL, Volkman BF, Westler WM, Heckman MP, Wu JH (March 2001). "Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain". J. Mol. Biol. 307 (3): 745–53. doi:10.1006/jmbi.2001.4522. PMID 11273698.
- ^ PDB: 1OHZ; Carvalho, A. L.; Dias, F. M. V.; Prates, J. A. M.; Nagy, T.; Gilbert, H. J.; Davies, G. J.; Ferreira, L. M. A.; Romao, M. J.; Fontes, C. M. G. A. (2003). "Cellulosome assembly revealed by the crystal structure of the cohesin-dockerin complex". Proceedings of the National Academy of Sciences. 100 (24): 13809–13814. doi:10.1073/pnas.1936124100. PMC 283503. PMID 14623971.
- ^ InterPro: InterPro: IPR016134
- ^ Adams JJ, Webb BA, Spencer HL, Smith SP (February 2005). "Structural characterization of type II dockerin module from the cellulosome of Clostridium thermocellum: calcium-induced effects on conformation and target recognition". Biochemistry. 44 (6): 2173–82. doi:10.1021/bi048039u. PMID 15697243.
External links
[edit]Protein Structure:
- Lytle BL, Volkman BF, Westler WM, Heckman MP, Wu JH (2001). "Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain". J Mol Biol. 307 (3): 745–753. doi:10.1006/jmbi.2001.4522. PMID 11273698.
- Bayer EA, Shimon LJ, Shoham Y, Lamed R (1998). "Cellulosomes-structure and ultrastructure". J Struct Biol. 124 (2–3): 221–234. doi:10.1006/jsbi.1998.4065. PMID 10049808.
Specificity Characterization:
- Haimovitz R, Barak Y, Morag E, Voronov-Goldman M, Shoham Y, Lamed R, Bayer EA (2008). "Cohesin-dockerin microarray: Diverse specificities between two complementary families of interacting protein modules". Proteomics. 8 (5): 968–979. doi:10.1002/pmic.200700486. PMID 18219699. S2CID 206360888.
- Adams JJ, Webb BA, Spencer HL, Smith SP (2005). "Structural characterization of type II dockerin module from the cellulosome of Clostridium thermocellum: calcium-induced effects on conformation and target recognition". Biochemistry. 44 (6): 2173–2182. doi:10.1021/bi048039u. PMID 15697243.
- Jindou S, Soda A, Karita S, Kajino T, Béguin P, Wu JH, Inagaki M, Kimura T, Sakka K, Ohmiya K (2004). "Cohesin-dockerin interactions within and between Clostridium josui and Clostridium thermocellum: binding selectivity between cognate dockerin and cohesin domains and species specificity". J Biol Chem. 279 (11): 9867–9874. doi:10.1074/jbc.M308673200. PMID 14688277.
