Haplogroup R1b: Difference between revisions
Sardinianr1b (talk | contribs) Removed a possible case of POV. |
→Origin and dispersal: there has never been another theory published, nor any criticism of it, so I think we can make this sentence simple |
||
| Line 47: | Line 47: | ||
}} |
}} |
||
The point of origin of R1b is thought |
The point of origin of R1b is thought to lie in [[Eurasia]], most likely in [[Western Asia]].<ref name=Myres2010/> T. Karafet et al. estimated the age of R1, the parent of R1b, as 18,500 years before present.<ref name=Karafet08/> |
||
In 2000 Ornella Semino and colleagues argued that R1b was ancient in Europe, and spread north from an Iberian Ice Age refuge after the Last Glacial Maximum.<ref>O. Semino et al, The genetic legacy of paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective, ''Science'', vol. 290 (2000), pp. 1155-59.</ref>This is one of main theories concerning the subject at hand and was supported by a mid 2010 study of the haplogroup R1b1b2 y-chromosome across Europe <ref>{{cite|last1=Morelli|title=A Comparison of Y-Chromosome Variation in Sardinia and Anatolia Is More Consistent with Cultural Rather than Demic Diffusion of Agriculture|year=2010|url=http://www.plosone.org/article/info:doi%2F10.1371%2Fjournal.pone.0010419|doi=10.1371/journal.pone.0010419|journal=PLoS ONE|volume=5|issue=4}}</ref> |
In 2000 Ornella Semino and colleagues argued that R1b was ancient in Europe, and spread north from an Iberian Ice Age refuge after the Last Glacial Maximum.<ref>O. Semino et al, The genetic legacy of paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective, ''Science'', vol. 290 (2000), pp. 1155-59.</ref>This is one of main theories concerning the subject at hand and was supported by a mid 2010 study of the haplogroup R1b1b2 y-chromosome across Europe <ref>{{cite|last1=Morelli|title=A Comparison of Y-Chromosome Variation in Sardinia and Anatolia Is More Consistent with Cultural Rather than Demic Diffusion of Agriculture|year=2010|url=http://www.plosone.org/article/info:doi%2F10.1371%2Fjournal.pone.0010419|doi=10.1371/journal.pone.0010419|journal=PLoS ONE|volume=5|issue=4}}</ref> |
||
Revision as of 09:30, 28 August 2010
| Haplogroup R1b | |
|---|---|
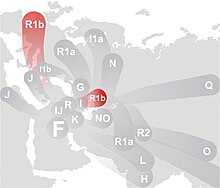 | |
| Possible time of origin | less than 18,500 years BP[1] |
| Possible place of origin | Southwest Asia [2] |
| Ancestor | R1 |
| Descendants | R1b1a (R-V88), R1b1b (R-P297) |
| Defining mutations | 1. M343 defines R1b in the broadest sense 2. P25 defines R1b1, making up most of R1b, and is often used to test for R1b 3. In some cases, major downstream mutations such as M269 are used to identify R1b, especially in regional or out-of-date studies |
| Highest frequencies | Western Europe, Northern Cameroon, Hazara, Bashkirs |

In human genetics, Haplogroup R1b is the most frequently occurring Y-chromosome haplogroup in Western Europe, parts of central Eurasia (for example Bashkortostan[3]), and in parts of sub-Saharan Central Africa (for example around Chad and Cameroon). R1b is also present at lower frequencies throughout Eastern Europe, Western Asia, Central Asia, and parts of North Africa. Due to European emigration it also reaches high frequencies in the Americas and Australia. While Western Europe is dominated by the R1b1b2 (R-M269) branch of R1b, the Chadic-speaking area in Africa is dominated by the branch known as R1b1a (R-V88). These represent two very successful "twigs" on a much bigger "family tree".
Nomenclature
"R1b", "R1b1", and so on are "phylogenetic" or family tree based names which explain the branching of the family tree of R1b. For example R1b1a and R1b1b would be branches of R1b1, descending from a common ancestor. This means that these names can change with new discoveries.
The alternative way of naming haplogroups is to refer to the mutations used to define and identify them, for example "R-M343" which is equivalent to "R1b". Haplogroup R1b is in other words now identified by the presence of the single-nucleotide polymorphism (SNP) mutation M343, which was discovered in 2004.[4] From 2002 to 2005, R1b was defined by the presence of SNP P25.
Standardized naming as described above, both using phylogenetic or mutational systems, was first proposed in 2002 by the Y Chromosome Consortium. Prior to 2002, today's Haplogroup R1b had a number of names in differing nomenclature systems, such as Hg1 and Eu18.[5]
After 2002, a major update of the YCC phylogenetic nomenclature was made in 2008 by Karafet et al. which took account of newer discoveries of branches which could be clearly defined by SNP mutations, including some which changed the understanding of R1b's family tree.[1] Since 2008 it has become increasing necessary to refer to the frequently updated listing made on the ISOGG website.[2]
Origin and dispersal
R1b is a sub-clade within the much larger Eurasian MNOPS "macro-haplogroup", which is one of the predominant groupings of all human male lines outside of Africa, and this whole group, along indeed with all of macro-haplogroup F, is believed to have originated in Asia.
| Macro-haplogroup MNOPS |
| ||||||||||||||||||||||||||||||||||||
The point of origin of R1b is thought to lie in Eurasia, most likely in Western Asia.[6] T. Karafet et al. estimated the age of R1, the parent of R1b, as 18,500 years before present.[1]
In 2000 Ornella Semino and colleagues argued that R1b was ancient in Europe, and spread north from an Iberian Ice Age refuge after the Last Glacial Maximum.[7]This is one of main theories concerning the subject at hand and was supported by a mid 2010 study of the haplogroup R1b1b2 y-chromosome across Europe [8]
Barbara Arredi and colleagues were the first to point out that the distribution of R1b forms a cline from east to west, which is far more consistent with an entry into Europe from Western Asia with the spread of farming.[9] Their conclusions on the eastern origin of R1b were supported by two detailed studies based on large datasets published in 2010. Both detected that the earliest subclades of R1b are found in western Asia and the most recent in western Europe.[10] Another paper added to this perspective by using R1b as an example of a wave haplogroup distribution, in this case from east to west.[11] However Myres et al. (August 2010) remained undecided on the dating of the migration or migrations responsible for this distribution. While concentrating their attention on the Neolithic, they do not rule out earlier or later movements.[6]
Root of R1b tree
The identifiers below are those from the 2010 revision of the ISOGG tree.[2]
| M343 |
| ||||||||||||||||||||||||||||||
R1b*
R1b* (that is R1b with no subsequent distinguishing SNP mutations) is extremely rare. Two cases were reported in a large study of Turkey.[4] In a study of Jordan it was found that no less than 20 out of all 146 men tested (13.7%), including most notably 20 out of 45 men tested from the Dead Sea area, were positive for M173 (R1) but negative for P25 and M269, mentioned above, as well as the R1a markers SRY10831.2 and M17, so they are either R1b* or R1a*.[12] Hassan et al. (2008) found an equally surprising 14 out of 26 (54%) of Sudanese Fulani who were M173+ and P25-.<[13] Wood et al. report 2 Egyptian cases of R1-M173 which were negative for SRY10831 (R1a1) and P25 (R1b1), out of a sample of 1122 males from various African countries, including 92 from Egypt.[14] Such cases could possibly be either R1b* (R-M343*) or R1a* (R-M420*) (demonstrating the importance of checking exact mutations tested when comparing findings in this field).
It is however also possible that some of the rare examples represent a reversion of marker P25 from a positive back to a negative ancestral state.[15]
Ancient clades within R1b1 (R-P25)
An up-to-date compilation of data taking the latest information into account can be found in Cruciani et al. (2010) which can be summarised as follows[16]. As will be discussed below however, in some parts of western and northwestern Europe, R-M269 frequencies can reach even higher levels.
| Continent | Population | #No. | Total% | R1b1* (R-P25*) | R1b1a (R-V88) | R1b1b2 (R-M269) | R1b1b1 (R-M73) |
| Africa | Northern Africa | 691 | 5.9% | 0.0% | 5.2% | 0.7% | 0.0% |
| Africa | Central Sahel Region | 461 | 23.0% | 0.0% | 23.0% | 0.0% | 0.0% |
| Africa | Western Africa | 123 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Africa | Eastern Africa | 442 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Africa | Southern Africa | 105 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Europe | Western Europeans | 465 | 57.8% | 0.0% | 0.0% | 57.8% | 0.0% |
| Europe | North western Europeans | 43 | 55.8% | 0.0% | 0.0% | 55.8% | 0.0% |
| Europe | Central Europeans | 77 | 42.9% | 0.0% | 0.0% | 42.9% | 0.0% |
| Europe | Italians | 1173 | 26.6% | 0.0% | 0.2% | 26.4% | 0.0% |
| Europe | Corsicans | 141 | 48.9% | 0.0% | 0.7% | 48.2% | 0.0% |
| Europe | North Eastern Europeans | 74 | 1.4% | 0.0% | 0.0% | 1.4% | 0.0% |
| Europe | Russians | 60 | 6.7% | 0.0% | 0.0% | 6.7% | 0.0% |
| Europe | Eastern Europeans | 149 | 20.8% | 0.0% | 0.0% | 20.8% | 0.0% |
| Europe | Balkanians | 510 | 13.1% | 0.0% | 0.2% | 12.9% | 0.0% |
| Asia | Western Asians | 328 | 5.8% | 0.0% | 0.3% | 5.5% | 0.0% |
| Asia | Southern Asians | 288 | 4.8% | 0.0% | 0.0% | 1.7% | 3.1% |
| Asia | South eastern Asians | 10 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Asia | North eastern Asians | 30 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Asia | Eastern Asians | 156 | 0.6% | 0.0% | 0.0% | 0.6% | 0.0% |
| TOTAL | 5326 |
R1b1*
R1b1*, like R1b* is rare. As mentioned above, examples are described in older articles, for example two in a sample from Turkey,[4] but most cases, especially in Africa, are now thought to be almost mostly in the more recently discovered sub-clade R-V88 (see below). Most or all examples of R1b therefore fall into subclades R1b1a (R-V88) or R1b1b (R-P297). Cruciani et al. in the large 2010 study found 3 cases amongst 1173 Italians, 1 out of 328 West Asians and 1 out of 156 East Asians.[16] Varzari found 3 cases in the Ukraine, in a study of 322 people from the Dniester-Carpathian region, who were P25 positive, but M269 negative.[17] Cases from older studies are mainly from Africa, the Middle East or Mediterranean, and are discussed below as probable cases of R1b1a (R-V88).
R1b1a
R1b1a is defined by the presence of SNP marker V88, the discovery of which was announced in 2010 by Cruciani et al.[16] Apart from individuals in southern Europe and Western Asia, the majority of R-V88 was found in northern and central Africa:
| Region | Population | Country | Language | N | Total% | R1b1a (R-V88) | R1b1b2 (R-M269) | R1b1a* (R-V88*) | R1b1a4 (R-V69) |
| N Africa | Composite | Morocco | AA | 338 | 0.0% | 0.3% | 0.6% | 0.3% | 0.0% |
| N Africa | Mozabite Berbers | Algeria | AA/Berber | 67 | 3.0% | 3.0% | 0.0% | 3.0% | 0.0% |
| N Africa | Northern Egyptians | Egypt | AA/Semitic | 49 | 6.1% | 4.1% | 2.0% | 4.1% | 0.0% |
| N Africa | Berbers from Siwa | Egypt | AA/Berber | 93 | 28.0% | 26.9% | 1.1% | 23.7% | 3.2% |
| N Africa | Baharia | Egypt | AA/Semitic | 41 | 7.3% | 4.9% | 2.4% | 0.0% | 4.9% |
| N Africa | Gurna Oasis | Egypt | AA/Semitic | 34 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| N Africa | Southern Egyptians | Egypt | AA/Semitic | 69 | 5.8% | 5.8% | 0.0% | 2.9% | 2.9% |
| C Africa | Songhai | Niger | NS/Songhai | 10 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| C Africa | Fulbe | Niger | NC/Atlantic | 7 | 14.3% | 14.3% | 0.0% | 14.3% | 0.0% |
| C Africa | Tuareg | Niger | AA/Berber | 22 | 4.5% | 4.5% | 0.0% | 4.5% | 0.0% |
| C Africa | Ngambai | Chad | NS/Sudanic | 11 | 9.1% | 9.1% | 0.0% | 9.1% | 0.0% |
| C Africa | Hausa | Nigeria (North) | AA/Chadic | 10 | 20.0% | 20.0% | 0.0% | 20.0% | 0.0% |
| C Africa | Fulbe | Nigeria (North) | NC/Atlantic | 32 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| C Africa | Yorubad | Nigeria (South) | NC/Defoid | 21 | 4.8% | 4.8% | 0.0% | 4.8% | 0.0% |
| C Africa | Ouldeme | Cameroon (Nth) | AA/Chadic | 22 | 95.5% | 95.5% | 0.0% | 95.5% | 0.0% |
| C Africa | Mada | Cameroon (Nth) | AA/Chadic | 17 | 82.4% | 82.4% | 0.0% | 76.5% | 5.9% |
| C Africa | Mafa | Cameroon (Nth) | AA/Chadic | 8 | 87.5% | 87.5% | 0.0% | 25.0% | 62.5% |
| C Africa | Guiziga | Cameroon (Nth) | AA/Chadic | 9 | 77.8% | 77.8% | 0.0% | 22.2% | 55.6% |
| C Africa | Daba | Cameroon (Nth) | AA/Chadic | 19 | 42.1% | 42.1% | 0.0% | 36.8% | 5.3% |
| C Africa | Guidar | Cameroon (Nth) | AA/Chadic | 9 | 66.7% | 66.7% | 0.0% | 22.2% | 44.4% |
| C Africa | Massa | Cameroon (Nth) | AA/Chadic | 7 | 28.6% | 28.6% | 0.0% | 14.3% | 14.3% |
| C Africa | Other Chadic | Cameroon (Nth) | AA/Chadic | 4 | 75.0% | 75.0% | 0.0% | 25.0% | 50.0% |
| C Africa | Shuwa Arabs | Cameroon (Nth) | AA/Semitic | 5 | 40.0% | 40.0% | 0.0% | 40.0% | 0.0% |
| C Africa | Kanuri | Cameroon (Nth) | NS/Saharan | 7 | 14.3% | 14.3% | 0.0% | 14.3% | 0.0% |
| C Africa | Foulbe | Cameroon (Nth) | NC/Atlantic | 18 | 11.1% | 11.1% | 0.0% | 5.6% | 5.6% |
| C Africa | Moundang | Cameroon (Nth) | NC/Adamawa | 21 | 66.7% | 66.7% | 0.0% | 14.3% | 52.4% |
| C Africa | Fali | Cameroon (Nth) | NC/Adamawa | 48 | 20.8% | 20.8% | 0.0% | 10.4% | 10.4% |
| C Africa | Tali | Cameroon (Nth) | NC/Adamawa | 22 | 9.1% | 9.1% | 0.0% | 4.5% | 4.5% |
| C Africa | Mboum | Cameroon (Nth) | NC/Adamawa | 9 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| C Africa | Composite | Cameroon (Sth) | NC/Bantu | 90 | 0.0% | 1.1% | 0.0% | 1.1% | 0.0% |
| C Africa | Biaka Pygmies | CAR | NC/Bantu | 33 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| W Africa | Composite | — | 123 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| E Africa | Composite | — | 442 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| S Africa | Composite | — | 105 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| TOTAL | 1822 |
| M343 |
| ||||||||||||||||||||||||||||||
As can be seen in the above data table, R1b1a is found in northern Cameroon in west central Africa at a very high frequency, where it is considered to be caused by a pre-Islamic movement of people from Eurasia.[14][18]
Suggestive results from other studies which did not test for the full range of new markers discovered by Cruciani et al. have also been reported, which might be in R-V88.
- Wood et al. reported high frequencies of men who were P25 positive and M269 negative, amongst the same north Cameroon area where Cruciani et al. reported high R-V88 levels. However they also found such cases amongst 3% (1/32) of Fante from Ghana, 9% (1/11) of Bassa from southern Cameroon, 4% (1/24) of Herero from Namibia, 5% (1/22) of Ambo from Namibia, 4% (4/92) of Egyptians, and 4% (1/28) of Tunisians.[14]
- Luis et al. found the following cases of men M173 positive (R1), but negative for M73 (R1b1b1), M269 (R1b1b2), M18 (R1b1a1, a clade with V88, M18 having been discovered before V88) and M17 (R1a1a): 1 of 121 Omanis, 3 of 147 Egyptians, 2 of 14 Bantu from southern Cameroon, and 1 of 69 Hutu from Rwanda.[19]
R1b1a1
R1b1a1 is a sub-clade of R-V88 which is defined by the presence of SNP marker M18.[1] It has been found only at low frequencies in samples from Sardinia[20][21] and Lebanon.[22]
R1b1b
R1b1b is defined by the presence of SNP marker P297. In 2008 this polymorphism was recognised to combine M73 and M269 into one R1b1b cluster.[1] The majority of Eurasian R1b is within this clade, representing a very large modern population. Although P297 itself has not yet been much tested for, the same population has been relatively well studied in terms of other markers. Therefore the branching within this clade can be explained in relatively high detail below.
R1b1c
R1b1c is defined by the presence of SNP marker M335. This haplogroup was created by the 2008 reorganisation of nomenclature and should not be confused with R1b1b2, which was previously called R1b1c. Its position in relation to the much more populous sub-clade R1b1b is uncertain.[1] The M335 marker was first published in 2004, when one example was discovered in Turkey, which was classified at that time as R1b4.[4]
Major clades within R1b1b (R-P297)
R1b1b1 (R-M73)
R1b1b1 is defined by the presence of SNP marker M73. It has been found at generally low frequencies throughout central Eurasia,[20] but has been found with relatively high frequency among particular populations there including Hazaras in Pakistan (8/25 = 32%)[23]; and Bashkirs in Bashkortostan (62/471 = 13.2%), 44 of these being found among the 80 tested Bashkirs of the Abzelilovsky District in the Republic of Bashkortostan (55.0%).[3] High levels were also spotted in very small samples from particular ethnic groups in China in the study by Sengupta et al in 2006. This included the Naxi (4/8) and Uygur (3/8), but R-M73 individuals were also found in the small samples for Japan, and the Chinese Mongola, Tu, and Han.[23] Four R-M73 men were also found in a 523 man study of Turkey.[4]
In 2010, Myres et al. report that out of 193 R-M173 men found amongst 10355 widespread men, "all except two Russians occurred outside Europe, either in the Caucasus, Turkey, the Circum-Uralic and North Pakistan regions".[24]
R1b1b2 (R-M269)
R1b1b2 is defined by the presence of SNP marker M269. European R1b is dominated by R-M269. It has been found at generally low frequencies throughout central Eurasia,[20] and with relatively high frequency among Bashkirs of the Bashkortostan and Perm region (84.0%).[3] Out of 523 men tested across Turkey 76 tested positive for M269[4].
| Long-hand: | R1b1b2 (formerly R1b1c, R1b3) |
| Defining SNP: | M269 |
| Parent Clade: | R-P297 |
| Subclades: | R-P311 |
From 2003 to 2005 what is now R1b1b2 was designated R1b3. From 2005 to 2008 it was R1b1c.
| M269 |
| |||||||||||||||||||||||||||
Within Europe, R-M269 is dominated by R-M412, otherwise known as R-L51, which according to Myres et al. (2010) is "virtually absent in the Near East, the Caucasus and West Asia". This Western European population is further divided between R-P312/S116 and R-U106/S21, which appear to spread from the western and eastern Rhine river basin respectively. Myres et al. note further that concerning it's closest relatives, in R-L23*, that it is "instructive" that these are often more than 10% of the population in the Caucasus, Turkey, and some southeast European and circum-Uralic populations. In Western Europe it is also present but in generally much lower levels apart from "an instance of 27% in Switzerland's Upper Rhone Valley".[6]
In articles published around 2000 it was proposed that this clade came into existence in Europe before the last Ice Age,[25] but more recently this scenario is no longer receiving much mainstream attention. A range of newer estimates for R1b1b2 are from 4,000 to a maximum of about 10,000 years ago, and coming from Western Asia via southeastern Europe.[2][9][6] Western European R1b is dominated by R-P310.[2] It is this Western European branch which is in turn dominated by U106 and P312, and the typical most common STR Y DNA signature for Western Europe, the so-called Atlantic Modal Haplotype, which is also sometimes referred to as "Haplotype 15".[2] "Haplotype 15" is contrasted with "Haplotype 35", which has long been noted as a distinct type of R1b1b2, more common towards the southeast of Europe. For example, in the Balkans, Georgia and Turkey "Haplotype 35" is prevalent[4]. In 2009, DNA extracted from the femur bones of 6 skeletons in an early-medieval burial place in Ergolding (Bavaria, Germany) dated to around 670 AD yielded the following results: 4 were found to be haplogroup R1b with the closest matches in modern populations of Germany. Ireland and the USA while 2 were in Haplogroup G2a.[26]
Population studies which test for M269 have become more common in recent years, while in earlier studies men in this haplogroup are only visible in the data by extrapolation of what is likely. The following gives a summary of most of the studies which specifically tested for M269, showing its distribution in Europe, North Africa, the Middle East and Central Asia as far as China and Nepal.
| Country | Sampling | sample | R-M269 | Source |
| Wales | National | 65 | 92.3% | Balaresque et al. (2009)[27] |
| Spain | Basques | 116 | 87.1% | Balaresque et al. (2009)[27] |
| Ireland | National | 796 | 85.4% | Moore et al. (2006)[28] |
| Spain | Catalonia | 80 | 81.3% | Balaresque et al. (2009)[27] |
| France | Ile et Vilaine | 82 | 80.5% | Balaresque et al. (2009)[27] |
| France | Haute-Garonne | 57 | 78.9% | Balaresque et al. (2009)[27] |
| England | Cornwall | 64 | 78.1% | Balaresque et al. (2009)[27] |
| France | Loire-Atlantique | 48 | 77.1% | Balaresque et al. (2009)[27] |
| France | Finistère | 75 | 76.0% | Balaresque et al. (2009)[27] |
| France | Basques | 61 | 75.4% | Balaresque et al. (2009)[27] |
| Spain | East Andalucia | 95 | 72.0% | Balaresque et al. (2009)[27] |
| Spain | Castilla La Mancha | 63 | 72.0% | Balaresque et al. (2009)[27] |
| France | Vendeé | 50 | 68.0% | Balaresque et al. (2009)[27] |
| France | Baie de Somme | 43 | 62.8% | Balaresque et al. (2009)[27] |
| England | Leicestershire | 43 | 62.0% | Balaresque et al. (2009)[27] |
| Italy | North-East (Ladin) | 79 | 60.8% | Balaresque et al. (2009)[27] |
| Spain | Galicia | 88 | 58.0% | Balaresque et al. (2009)[27] |
| Spain | West Andalucia | 72 | 55.0% | Balaresque et al. (2009)[27] |
| Portugal | South | 78 | 46.2% | Balaresque et al. (2009)[27] |
| Italy | North-West | 99 | 45.0% | Balaresque et al. (2009)[27] |
| Denmark | National | 56 | 42.9% | Balaresque et al. (2009)[27] |
| Netherlands | National | 84 | 42.0% | Balaresque et al. (2009)[27] |
| Italy | North East | 67 | 41.8% | Battaglia et al. (2008)[29] |
| Russia | Bashkirs | 471 | 34.40% | Lobov (2009)[3] |
| Germany | Bavaria | 80 | 32.3% | Balaresque et al. (2009)[27] |
| Italy | West Sicily | 122 | 30.3% | Di Gaetano et al. (2009)[30] |
| Slovenia | National | 75 | 21.3% | Battaglia et al. (2008)[29] |
| Slovenia | National | 70 | 20.6% | Balaresque et al. (2009)[27] |
| Turkey | Central | 152 | 19.1% | Cinnioğlu et al. (2004)[4] |
| Republic of Macedonia | Albanians | 64 | 18.8% | Battaglia et al. (2008)[29] |
| Italy | East Sicily | 114 | 18.4% | Di Gaetano et al. (2009)[30] |
| Crete | National | 193 | 17.0% | King et al. (2008)[31] |
| Italy | Sardinia | 930 | 17.0% | Contu et al (2008)[32] |
| Iran | North | 33 | 15.2% | Regueiro et al. (2006)[33] |
| Moldova | 268 | 14.6% | Varzari (2006)[17] | |
| Greece | National | 171 | 13.5% | King et al. (2008)[31] |
| Turkey | West | 163 | 13.5% | Cinnioğlu et al. (2004)[4] |
| Romania | National | 54 | 13.0% | Varzari (2006)[17] |
| Turkey | East | 208 | 12.0% | Cinnioğlu et al. (2004)[4] |
| Algeria | 93 | 11.8% | Robino et al. (2008)[34] | |
| Russia | Roslavl | 107 | 11.2% | Balanovsky et al. (2008)[35] |
| Iraq | National | 139 | 10.8% | Al-Zahery et al. (2003)[36] |
| Nepal | Newar | 66 | 10.60% | Gayden et al. (2007)[37] |
| Serbia | National | 100 | 10.0% | Belaresque et al. (2009)[27] |
| Bosnia-Herzegovina | Serb | 81 | 6.2% | Marjanovic et al. (2005)[38] |
| Iran | South | 117 | 6.0% | Regueiro et al. (2006)[33] |
| Russia | Repievka | 96 | 5.2% | Balanovsky et al. (2008)[39] |
| UAE | 164 | 3.7% | Cadenas et al. (2007)[40] | |
| Bosnia-Herzegovina | Bosniak | 85 | 3.5% | Marjanovic et al. (2005)[38] |
| Pakistan | 176 | 2.8% | Sengupta et al. (2006)[23] | |
| Russia | Belgorod | 143 | 2.8% | Balanovsky et al. (2008)[39] |
| Russia | Ostrov | 75 | 2.7% | Balanovsky et al. (2008)[39] |
| Russia | Pristen | 45 | 2.2% | Balanovsky et al. (2008)[39] |
| Bosnia-Herzegovina | Croat | 90 | 2.2% | Marjanovic et al. (2005)[38] |
| Qatar | 72 | 1.4% | Cadenas et al. (2007)[40] | |
| China | 128 | 0.8% | Sengupta et al. (2006)[23] | |
| India | various | 728 | 0.5% | Sengupta et al. (2006)[23] |
| Croatia | Osijek | 29 | 0.0% | Battaglia et al. (2008)[29] |
| Yemen | 62 | 0.0% | Cadenas et al. (2007)[40] | |
| Tibet | 156 | 0.0% | Gayden et al. (2007)[37] | |
| Nepal | Tamang | 45 | 0.0% | Gayden et al. (2007)[37] |
| Nepal | Kathmandu | 77 | 0.0% | Gayden et al. (2007)[37] |
| Japan | 23 | 0.0% | Sengupta et al. (2006)[23] |
R1b1b2a1a1 (R-U106) - a sub-clade within R1b1b2
This subclade is defined by the presence of the marker U106, also known as S21 and M405.[2][41] It appears to represent over 25% of R1b in Europe.[2]
| Phylogenetic name: | R1b1b2a1a1 |
| Defining SNP: | U106/S21/M405 |
| Parent Clade: | P310/S129 |
| Subclades: | U198/S29/M467, P107, S26/L1/DYS439(null), L48/S162 (comprising L44, L45, L46, L47, L148), L257 |
| U106/S21 |
| ||||||||||||||||||||||||||||||||||||
While this sub-clade of R1b is frequently discussed amongst genetic genealogists, the following table represents the peer-reviewed findings published so far in the 2007 articles of Myres et al. and Sims et al.[41][24]
| Population | Sample size | R-M269 | R-U106 (without U198) | R-U198 |
| Austria | 22 | 27.30% | 22.70% | 0.00% |
| Central/South America | 33 | 0.00% | 0.00% | 0.00% |
| Czech Republic | 36 | 27.80% | 13.90% | 0.00% |
| Denmark | 113 | 34.50% | 16.80% | 0.09% |
| Eastern Europe | 44 | 4.50% | 0.00% | 0.00% |
| England | 138 | 57.20% | 20.30% | 0.14% |
| France | 56 | 51.80% | 7.10% | 0.00% |
| Germany | 332 | 43.10% | 18.70% | 0.18% |
| Ireland | 102 | 80.40% | 5.90% | 0.00% |
| Italy | 284 | 37.30% | 3.50% | 0.00% |
| Jordan | 76 | 0.00% | 0.00% | 0.00% |
| Middle-East | 43 | 0.00% | 0.00% | 0.00% |
| Netherlands | 94 | 54.30% | 35.10% | 0.21% |
| Oceania | 43 | 0.00% | 0.00% | 0.00% |
| Oman | 29 | 0.00% | 0.00% | 0.00% |
| Pakistan | 177 | 3.40% | 0.00% | 0.00% |
| Palestine | 47 | 0.00% | 0.00% | 0.00% |
| Poland | 110 | 22.70% | 8.20% | 0.00% |
| Russia | 56 | 21.40% | 5.40% | 0.18% |
| Slovenia | 105 | 17.10% | 3.80% | 0.00% |
| Switzerland | 90 | 57.80% | 13.30% | 0.00% |
| Turkey | 523 | 14.50% | 0.40% | 0.00% |
| Ukraine | 32 | 25.00% | 9.40% | 0.00% |
| United States | 58 | 5.20% | 5.20% | 0.00% |
| US (European) | 125 | 46.40% | 14.40% | 0.80% |
| US (Afroamerican) | 118 | 14.30% | 1.70% | 0.80% |
R1b1b2a1a2 (R-P312) - a sub-clade within R1b1b2
Along with R-U106, R-P312 is one of the most common types of R1b1b2 (R-M269) in Europe. It has been the subject of significant study concerning its sub-clades, and those so far recognized by the ISOGG website are summarized in the following table.[2]
| Phylogenetic name: | R1b1b2a1a2 |
| Defining SNP: | P312 (also called S116, rs34276300) |
| Parent Clade: | R-P310 |
| Subclades: | R-M153, R-M167, R-U152, R-L21 |
| P312 |
| |||||||||||||||||||||||||||||||||||||||||||||
R1b1b2a1a2b
This subclade is defined by the presence of the marker R-M153. It has been found mostly in Basques and Gascons, among whom it represents a sizeable fraction of the Y-DNA pool[42][43], though is also found occasionally among Iberians in general. The first time it was located (Bosch 2001[44]) it was described as H102 and included 7 Basques and one Andalusian.
R1b1b2a1a2c
This subclade is defined by the presence of the marker M167, also known as SRY2627. The first author to test for this marker (long before current haplogroup nomenclature existed) was Hurles in 1999, who tested 1158 men in various populations[45]. He found it relatively common among Basques (13/117: 11%) and Catalans (7/32: 22%). Other occurrences were found among other Spanish, Béarnais, other French, British and Germans.
In 2000 Rosser et al., in a study which tested 3616 men in various populations[46] also tested for that same marker, naming the haplogroup Hg22, and again it was found mainly among Basques (19%), in lower frequencies among French (5%), Bavarians (3%), Spanish (2%), Southern Portuguese (2%), and in single occurrences among Romanians, Slovenians, Dutch, Belgians and English.
In 2001 Bosch described this marker as H103, in 5 Basques and 5 Catalans.[44] Further regional studies have located it in significant amounts in Asturias, Cantabria and Galicia, as well as again among Basques.[44] Cases in the Azores and Latin America have also been reported. In 2008 two research papers by López-Parra[43] and Adams,[42] respectively, confirmed a strong association with all or most of the Pyrenees and Eastern Iberia.
In a larger study of Portugal in 2006, with 657 men tested, Beleza et al. confirmed similar low levels in all the major regions, from 1.5%-3.5%.[47]
R1b1b2a1a2d
This subclade is defined by the presence of the marker U152, also called S28[2]. Its discovery was announced in 2005 by EthnoAncestry[48] and subsequently identified independently by Sims et al. (2007).[41] Out of a sample of 135 men in Tyrol, Austria, 45 men tested positive for M343 (R1b). Of these 45, 25 tested positive for U106/S21, and 9 for U152/S28. 8 Men could not be further identified in the study. One man testing positive for U106 also tested positive for U198[49]. Myres et al. report this clade "is most frequent (20-44%) in Switzerland, Italy, France and Western Poland, with additional instances exceeeding 15% in some regions of England and Germany".[24]
R1b1b2a1a2e
This subclade is defined by the presence of the marker S68, also known as L165.
R1b1b2a1a2f
This subclade is defined by the presence of the marker L21, also referred to as M529.[2] Myres et al. report it is most common in England and Ireland (25-50% of the whole male population).[6]
R1b1b2a1a2f2
This subclade within R-L21 is defined by the presence of the marker M222. It is particularly associated with male lines which are Irish or Scottish. In this case, the relatively high frequency of this specific subclade among the population of certain counties in northwestern Ireland may be due to positive social selection, as it is suggested to have been the Y-chromosome haplogroup of the Uí Néill dynastic kindred of ancient Ireland.[28] However it is not restricted to the Uí Néill as it is also associated with the closely related Connachta dynasties, called the Uí Briúin and Uí Fiachrach.[50] M222 is also found as a substantial proportion of the population of Scotland which may indicate substantial settlement from northern Ireland or at least links to it.[28][51] Those areas settled by large numbers of Irish and Scottish emigrants such as North America have a substantial percentage of M222.[28]
R1b1b2a1a2f4
This subclade is defined by the presence of the marker L226, also known as S168. Commonly referred to as Irish Type III, it is concentrated in central western Ireland and associated with the Dál gCais kindred.[52]
Popular culture
Bryan Sykes, in his book Blood of the Isles, gives the populations associated with R1b the name of Oisín for a clan patriarch, much as he did for mitochondrial haplogroups in The Seven Daughters of Eve.
Stephen Oppenheimer also deals with this haplogroup in his book Origins of the British, giving the R1b clan patriarch the Basque name "Ruisko" in honour of what he thought was the Iberian origin of R1b.
See also
- Human Y-chromosome DNA haplogroup
- Atlantic Modal Haplotype
- Genealogical DNA test
- Prehistoric Europe
- Y-DNA haplogroups by ethnic groups
References
- ^ a b c d e f Karafet, TM; Mendez, FL; Meilerman, MB; Underhill, PA; Zegura, SL; Hammer, MF (2008). "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome research. 18 (5): 830–8. doi:10.1101/gr.7172008. PMC 2336805. PMID 18385274.
- ^ a b c d e f g h i j k International Society of Genetic Genealogy (ISOGG) - Y-DNA Haplogroup R and its Subclades
- ^ a b c d A. S. Lobov et al. (2009), "Structure of the Gene Pool of Bashkir Subpopulations" (original text in Russian)
- ^ a b c d e f g h i j Cinnioğlu, C; King, R; Kivisild, T; Kalfoğlu, E; Atasoy, S; Cavalleri, GL; Lillie, AS; Roseman, CC; Lin, AA (2004). "Excavating Y-chromosome haplotype strata in Anatolia" (PDF). Human genetics. 114 (2): 127–48. doi:10.1007/s00439-003-1031-4. PMID 14586639.
- ^ Y Chromosome Consortium (18 January 2002). "YCC NRY Tree 2002". Retrieved 13 December 2007.
- ^ a b c d e Myres, Natalie (2010), "A major Y-chromosome haplogroup R1b Holocene effect in Central and Western Europe", European Journal of Human Genetics
- ^ O. Semino et al, The genetic legacy of paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective, Science, vol. 290 (2000), pp. 1155-59.
- ^ Morelli (2010), "A Comparison of Y-Chromosome Variation in Sardinia and Anatolia Is More Consistent with Cultural Rather than Demic Diffusion of Agriculture", PLoS ONE, 5 (4), doi:10.1371/journal.pone.0010419
{{citation}}: CS1 maint: unflagged free DOI (link) - ^ a b B. Arredi, E. S. Poloni and C. Tyler-Smith (2007). "The peopling of Europe". In Crawford, Michael H. (ed.). Anthropological genetics: theory, methods and applications. Cambridge, UK: Cambridge University Press. p. 394. ISBN 0-521-54697-4.
- ^ P.Balaresque et al., A predominantly neolithic origin for European paternal lineages, PLoS Biology, vol. 8, no. 1 (January 2010); N.M Myres et al., A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe, European Journal of Human Genetics, (advance online publication 25 August 2010).
- ^ J. Chiaroni, P. Underhill and L.L. Cavalli-Sforza, Y chromosome diversity, human expansion, drift and cultural evolution, Proceedings of the National Academy of Sciences of the United States of America, vol. 106, no. 48 (December 2009), pp. 20174-79.
- ^ Flores, C; Maca-Meyer, N; Larruga, JM; Cabrera, VM; Karadsheh, N; Gonzalez, AM (2005). "Isolates in a corridor of migrations: a high-resolution analysis of Y-chromosome variation in Jordan". Journal of human genetics. 50 (9): 435–41. doi:10.1007/s10038-005-0274-4. PMID 16142507.
- ^ Hassan, HY; Underhill, PA; Cavalli-Sforza, LL; Ibrahim, ME (2008). "Y-chromosome variation among Sudanese: restricted gene flow, concordance with language, geography, and history" (PDF). American journal of physical anthropology. 137 (3): 316–23. doi:10.1002/ajpa.20876. PMID 18618658.
13/32
- ^ a b c Wood, ET; Stover, DA; Ehret, C; Destro-Bisol, G; Spedini, G; Mcleod, H; Louie, L; Bamshad, M; Strassmann, BI (2005). "Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes" (PDF). European journal of human genetics : EJHG. 13 (7): 867–76. doi:10.1038/sj.ejhg.5201408. PMID 15856073.
- ^ Adams, SM; King, TE; Bosch, E; Jobling, MA (2006). "The case of the unreliable SNP: recurrent back-mutation of Y-chromosomal marker P25 through gene conversion". Forensic science international. 159 (1): 14–20. doi:10.1016/j.forsciint.2005.06.003. PMID 16026953.
- ^ a b c Cruciani; Trombetta, B; Sellitto, D; Massaia, A; Destro-Bisol, G; Watson, E; Beraud Colomb, E; Dugoujon, JM; Moral, P; et al. (2010). "Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages". European Journal of Human Genetics. 18 (7): 800–7. doi:10.1038/ejhg.2009.231. PMID 20051990.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ a b c Varzari, Alexander (2006), "Population History of the Dniester-Carpathians: Evidence from Alu Insertion and Y-Chromosome Polymorphisms" (PDF), Dissertation der Fakultät für Biologie der Ludwig-Maximilians-Universität München
- ^ Cruciani, F; Santolamazza, P; Shen, P; Macaulay, V; Moral, P; Olckers, A; Modiano, D; Holmes, S; Destro-Bisol, G (2002). "A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes". American journal of human genetics. 70 (5): 1197–214. doi:10.1086/340257. PMC 447595. PMID 11910562., pp. 13–14
- ^ Luis, JR; Rowold, DJ; Regueiro, M; Caeiro, B; Cinnioğlu, C; Roseman, C; Underhill, PA; Cavalli-Sforza, LL; Herrera, RJ (2004). "The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations". American journal of human genetics. 74 (3): 532–44. doi:10.1086/382286. PMC 1182266. PMID 14973781.
- ^ a b c Peter A. Underhill, Peidong Shen, Alice A. Lin et al., "Y chromosome sequence variation and the history of human populations", Nature Genetics, Volume 26, November 2000
- ^ Contu, D; Morelli, L; Santoni, F; Foster, JW; Francalacci, P; Cucca, F; Hawks, John (2008). "Y-chromosome based evidence for pre-neolithic origin of the genetically homogeneous but diverse Sardinian population: inference for association scans". PloS one. 3 (1): e1430. doi:10.1371/journal.pone.0001430. PMC 2174525. PMID 18183308.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Zalloua, PA; Xue, Y; Khalife, J; Makhoul, N; Debiane, L; Platt, DE; Royyuru, AK; Herrera, RJ; Hernanz, DF (2008). "Y-chromosomal diversity in Lebanon is structured by recent historical events". American journal of human genetics. 82 (4): 873–82. doi:10.1016/j.ajhg.2008.01.020. PMC 2427286. PMID 18374297.
- ^ a b c d e f Sengupta, S; Zhivotovsky, LA; King, R; Mehdi, SQ; Edmonds, CA; Chow, CE; Lin, AA; Mitra, M; Sil, SK (2006). "Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists". American journal of human genetics. 78 (2): 202–21. doi:10.1086/499411. PMC 1380230. PMID 16400607.
8/176 R-M73 and 5/176 R-M269 for a total of 13/176 R1b in Pakistan and 4/728 R-M269 in India
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b c Myres, NM; Ekins, JE; Lin, AA; Cavalli-Sforza, LL; Woodward, SR; Underhill, PA (2007). "Y-chromosome short tandem repeat DYS458.2 non-consensus alleles occur independently in both binary haplogroups J1-M267 and R1b3-M405". Croatian medical journal. 48 (4): 450–9. PMC 2080563. PMID 17696299.
- ^ Semino, O; Passarino, G; Oefner, PJ; Lin, AA; Arbuzova, S; Beckman, LE; De Benedictis, G; Francalacci, P; Kouvatsi, A (2000). "The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective". Science. 290 (5494): 1155–9. doi:10.1126/science.290.5494.1155. PMID 11073453.
- ^ Vanek, Daniel (2009). "Kinship and Y-Chromosome Analysis of 7th Century Human Remains: Novel DNA Extraction and Typing Procedure for Ancient Material". Croatian Medical Journal. 3. 50: 286–295.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b c d e f g h i j k l m n o p q r s t u v w x Balaresque; et al. (2010), "A Predominantly Neolithic Origin for European Paternal Lineages", PLoS Biol., 8 (1), doi:10.1371/journal.pbio.1000285, PMID PMC2799514
{{citation}}: Check|pmid=value (help); Explicit use of et al. in:|author=(help)CS1 maint: unflagged free DOI (link) - ^ a b c d Moore; McEvoy, B; Cape, E; Simms, K; Bradley, DG; et al. (2006). "A Y-Chromosome Signature of Hegemony in Gaelic Ireland". American journal of human genetics. 78 (2): 334–8. doi:10.1086/500055. PMC 1380239. PMID 16358217.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ a b c d Battaglia, V; Fornarino, S; Al-Zahery, N; Olivieri, A; Pala, M; Myres, NM; King, RJ; Rootsi, S; Marjanovic, D (2009). "Y-chromosomal evidence of the cultural diffusion of agriculture in Southeast Europe". European journal of human genetics : EJHG. 17 (6): 820–30. doi:10.1038/ejhg.2008.249. PMID 19107149.
- ^ a b Di Gaetano, C; Cerutti, N; Crobu, F; Robino, C; Inturri, S; Gino, S; Guarrera, S; Underhill, PA; King, RJ (2009). "Differential Greek and northern African migrations to Sicily are supported by genetic evidence from the Y chromosome". European journal of human genetics : EJHG. 17 (1): 91–9. doi:10.1038/ejhg.2008.120. PMID 18685561.
57/232
- ^ a b King, RJ; Ozcan, SS; Carter, T; Kalfoğlu, E; Atasoy, S; Triantaphyllidis, C; Kouvatsi, A; Lin, AA; Chow, CE (2008). "Differential Y-chromosome Anatolian influences on the Greek and Cretan Neolithic". Annals of human genetics. 72 (Pt 2): 205–14. doi:10.1111/j.1469-1809.2007.00414.x. PMID 18269686.
- ^ Contu, D; Morelli, L; Santoni, F; Foster, JW; Francalacci, P; Cucca, F; Hawks, John (2008). "Y-chromosome based evidence for pre-neolithic origin of the genetically homogeneous but diverse Sardinian population: inference for association scans". PloS one. 3 (1): e1430. doi:10.1371/journal.pone.0001430. PMC 2174525. PMID 18183308.
174/930
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Regueiro, M; Cadenas, AM; Gayden, T; Underhill, PA; Herrera, RJ; et al. (2006). "Iran: Tricontinental Nexus for Y-Chromosome Driven Migration" (PDF). Hum Hered. 61 (3): 132–143. doi:10.1159/000093774. PMID 16770078.
{{cite journal}}: Explicit use of et al. in:|last=(help) - ^ Robino; et al. (2008), "Analysis of Y-chromosomal SNP haplogroups and STR haplotypes in an Algerian population sample", Journal International Journal of Legal Medicine, 122 (3): 251, doi:10.1007/s00414-007-0203-5
{{citation}}: Explicit use of et al. in:|author=(help) - ^ Balaresque; et al. (2010), "A Predominantly Neolithic Origin for European Paternal Lineages", PLoS Biol., 8 (1), doi:10.1371/journal.pbio.1000285, PMID PMC2799514
{{citation}}: Check|pmid=value (help); Explicit use of et al. in:|author=(help)CS1 maint: unflagged free DOI (link) - ^ Al-Zahery, N; Semino, O; Benuzzi, G; Magri, C; Passarino, G; Torroni, A; Santachiara-Benerecetti, AS (2003). "Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations" (PDF). Molecular phylogenetics and evolution. 28 (3): 458–72. doi:10.1016/S1055-7903(03)00039-3. PMID 12927131.
16/139
- ^ a b c d Gayden, T; Cadenas, AM; Regueiro, M; Singh, NB; Zhivotovsky, LA; Underhill, PA; Cavalli-Sforza, LL; Herrera, RJ (2007). "The Himalayas as a directional barrier to gene flow". American journal of human genetics. 80 (5): 884–94. doi:10.1086/516757. PMC 1852741. PMID 17436243.
- ^ a b c Marjanovic D, Fornarino S, Montagna S; et al. (2005). "The peopling of modern Bosnia-Herzegovina: Y-chromosome haplogroups in the three main ethnic groups". Ann. Hum. Genet. 69 (Pt 6): 757–63. doi:10.1111/j.1529-8817.2005.00190.x. PMID 16266413.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b c d Balanovsky; et al. (2008), "Two Sources of the Russian Patrilineal Heritage in Their Eurasian Context", AJHG, 82 (1): 236–250, doi:10.1016/j.ajhg.2007.09.019
{{citation}}: Explicit use of et al. in:|last=(help) - ^ a b c Cadenas; et al. (2007), "Y-chromosome diversity characterizes the Gulf of Oman", European Journal of Human Genetics, 16: 1–13, doi:10.1038/sj.ejhg.5201934
{{citation}}: Explicit use of et al. in:|author=(help) - ^ a b c Sims, LM; Garvey, D; Ballantyne, J (2007). "Sub-populations within the major European and African derived haplogroups R1b3 and E3a are differentiated by previously phylogenetically undefined Y-SNPs" (PDF). Human mutation. 28 (1): 97. doi:10.1002/humu.9469. PMID 17154278.
- ^ a b Adams, SM; Bosch, E; Balaresque, PL; Ballereau, SJ; Lee, AC; Arroyo, E; López-Parra, AM; Aler, M; Grifo, MS (2008). "The genetic legacy of religious diversity and intolerance: paternal lineages of Christians, Jews, and Muslims in the Iberian Peninsula". American journal of human genetics. 83 (6): 725–36. doi:10.1016/j.ajhg.2008.11.007. PMC 2668061. PMID 19061982.
- ^ a b López-Parra, AM; Gusmão, L; Tavares, L; Baeza, C; Amorim, A; Mesa, MS; Prata, MJ; Arroyo-Pardo, E (2009). "In search of the pre- and post-neolithic genetic substrates in Iberia: evidence from Y-chromosome in Pyrenean populations". Annals of human genetics. 73 (1): 42–53. doi:10.1111/j.1469-1809.2008.00478.x. PMID 18803634.
- ^ a b c Bosch, E; Calafell, F; Comas, D; Oefner, PJ; Underhill, PA; Bertranpetit, J (2001). "High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula". American journal of human genetics. 68 (4): 1019–29. doi:10.1086/319521. PMC 1275654. PMID 11254456.
- ^ Hurles, ME; Veitia, R; Arroyo, E; Armenteros, M; Bertranpetit, J; Pérez-Lezaun, A; Bosch, E; Shlumukova, M; Cambon-Thomsen, A (1999). "Recent male-mediated gene flow over a linguistic barrier in Iberia, suggested by analysis of a Y-chromosomal DNA polymorphism". American journal of human genetics. 65 (5): 1437–48. doi:10.1086/302617. PMC 1288297. PMID 10521311.
- ^ Rosser, ZH; Zerjal, T; Hurles, ME; Adojaan, M; Alavantic, D; Amorim, A; Amos, W; Armenteros, M; Arroyo, E (2000). "Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language". American Journal of Human Genetics. 67 (6): 1526–43. doi:10.1086/316890. PMC 1287948. PMID 11078479.
- ^ Beleza, S; Gusmão, L; Lopes, A; Alves, C; Gomes, I; Giouzeli, M; Calafell, F; Carracedo, A; Amorim, A (2006). "Micro-phylogeographic and demographic history of Portuguese male lineages". Annals of human genetics. 70 (Pt 2): 181–94. doi:10.1111/j.1529-8817.2005.00221.x. PMID 16626329.
395/657
- ^ http://ethnoancestry.com/R1b.html[unreliable source?]
- ^ Niederstätter, Harald, Recently introduced Y-SNPs improve the resolution within Y-chromosome haplogroup R1b in a central European population sample (Tyrol, Austria)
- ^ O'Neill; McLaughlin (2006.), "Insights Into the O'Neills of Ireland from DNA Testing", Journal of Genetic Genealogy
{{citation}}: Check date values in:|year=(help)CS1 maint: year (link) - ^ Campbell, Kevin D. (2007). "Geographic Patterns of Haplogroup R1b in the British Isles" (PDF). Journal of Genetic Genealogy. 3: 1–13.
- ^ Wright (2009). "A Set of Distinctive Marker Values Defines a Y-STR Signature for Gaelic Dalcassian Families". Journal of Genetic Genealogy.
External links
- International Society of Genetic Genealogy (ISOGG) - Y-DNA Haplogroup R and its Subclades - 2009
- EthnoAncestry R1b info/ Map of S21 and S29 frequencies as determined by EthnoAncestry's research arm
- Spread of R1b, from the Genographic Project, National Geographic
- R1b1b2 Subclade Maps: M167, M222, S21, S28
- World Haplogroup Maps Note especially the dominance of R1b in Western Europe.
- General SNP Marker Page DNA Heritage
- John McEwan's Y Chromosome DNA Genealogy Page
- David Faux - Shetland Isles Haplogroup R1b
- Dennis Wright - Irish Type III Homepage
Maps
Projects
- R1* Haplogroup Project
- R-ht35 DNA Project
- FTDNA Kerchner's R1b and Subclades YDNA Haplogroup Project
- R-P312 and Subclades Project
- R1b1b1 Haplogroup Project
- R1b1b2* Haplogroup Project
- R1b-U106/S21+ aka R1b1b2a1a Haplogroup Project
- R1b1b2a1b4 aka R1b(U152+) Project
- R1b1b2a1b5 aka R1b1b2a1a2f aka L21+ Project
- R1b1b2a1b5 aka R1b1b2a1a2f aka L21+ "Walk the Y" Project
- R1b-U198/S29+ Project
- R-M222 Haplogroup Project
- R-L226 "Irish Type III" Project
- R1b1* DNA Project
- null439 project
- British Isles DNA Project
- Wales Cymru DNA Project
- Danish Demes Regional DNA Project: Haplogroup R1b
- Jewish DNA Project
- FTDNA The Jewish R1b Project
- French Heritage DNA Project
- Iberian Peninsula DNA Project
- Normandy Y-DNA Project
- Scandinavia Y-DNA Project
- Norway DNA Project
- Benelux DNA Project
- Bretagne DNA Project
- Assyrian Heritage DNA Project
- Greek DNA Project
- Turkey DNA Project
- Czech DNA Project
- Switzerland DNA Project
- Polish Family Tree DNA Project
- Russia DNA Project
- Slovakia DNA Project
- Hungarian Magyar Project
- North Italy DNA Project
- Italy DNA Project
- Sicily DNA Project
