Conosa: Difference between revisions
added phylogeny section |
added tree diagram which was adapted from the Lobosa page |
||
| Line 30: | Line 30: | ||
While morphological characteristics like pseudopodia and body shape, flagella, and cytoplasm properties have not been regarded as convincing taxonomic suggestions<ref name="pmid118306644">{{cite journal |display-authors=etal |vauthors=Bapteste E, Brinkmann H, Lee JA |date=February 2002 |title=The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba |journal=Proc. Natl. Acad. Sci. U.S.A. |volume=99 |issue=3 |pages=1414–9 |bibcode=2002PNAS...99.1414B |doi=10.1073/pnas.032662799 |pmc=122205 |pmid=11830664 |doi-access=free}}</ref>, emerging sequencing data is being used to support Conosa’s monophyly. A study using several hundred phylogenetic markers of 30 species found Conosa to be monophyletic as representatives of Mycetozoa, Entamoebidae, and Pelobionta grouped together using several amino acid sequencing analysis methods.<ref name="pmid118306644" /> The monophyly of Conosa and the Archamoebea infraphyla was also supported by cDNA sequencing of 17 Amoebozoans.<ref name=":13">{{Cite journal |last=Cavalier-Smith |first=Thomas |last2=Fiore-Donno |first2=Anna Maria |last3=Chao |first3=Ema |last4=Kudryavtsev |first4=Alexander |last5=Berney |first5=Cédric |last6=Snell |first6=Elizabeth A. |last7=Lewis |first7=Rhodri |date=2015-02 |title=Multigene phylogeny resolves deep branching of Amoebozoa |url=https://linkinghub.elsevier.com/retrieve/pii/S1055790314002784 |journal=Molecular Phylogenetics and Evolution |language=en |volume=83 |pages=293–304 |doi=10.1016/j.ympev.2014.08.011}}</ref> |
While morphological characteristics like pseudopodia and body shape, flagella, and cytoplasm properties have not been regarded as convincing taxonomic suggestions<ref name="pmid118306644">{{cite journal |display-authors=etal |vauthors=Bapteste E, Brinkmann H, Lee JA |date=February 2002 |title=The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba |journal=Proc. Natl. Acad. Sci. U.S.A. |volume=99 |issue=3 |pages=1414–9 |bibcode=2002PNAS...99.1414B |doi=10.1073/pnas.032662799 |pmc=122205 |pmid=11830664 |doi-access=free}}</ref>, emerging sequencing data is being used to support Conosa’s monophyly. A study using several hundred phylogenetic markers of 30 species found Conosa to be monophyletic as representatives of Mycetozoa, Entamoebidae, and Pelobionta grouped together using several amino acid sequencing analysis methods.<ref name="pmid118306644" /> The monophyly of Conosa and the Archamoebea infraphyla was also supported by cDNA sequencing of 17 Amoebozoans.<ref name=":13">{{Cite journal |last=Cavalier-Smith |first=Thomas |last2=Fiore-Donno |first2=Anna Maria |last3=Chao |first3=Ema |last4=Kudryavtsev |first4=Alexander |last5=Berney |first5=Cédric |last6=Snell |first6=Elizabeth A. |last7=Lewis |first7=Rhodri |date=2015-02 |title=Multigene phylogeny resolves deep branching of Amoebozoa |url=https://linkinghub.elsevier.com/retrieve/pii/S1055790314002784 |journal=Molecular Phylogenetics and Evolution |language=en |volume=83 |pages=293–304 |doi=10.1016/j.ympev.2014.08.011}}</ref> |
||
The last common ancestor of Conosa was likely an aerobic [[protist]] with anterior and recurrent flagellum.<ref name=":03">{{Cite journal |last=Pánek |first=Tomáš |last2=Zadrobílková |first2=Eliška |last3=Walker |first3=Giselle |last4=Brown |first4=Matthew W. |last5=Gentekaki |first5=Eleni |last6=Hroudová |first6=Miluše |last7=Kang |first7=Seungho |last8=Roger |first8=Andrew J. |last9=Tice |first9=Alexander K. |last10=Vlček |first10=Čestmír |last11=Čepička |first11=Ivan |date=2016-05-01 |title=First multigene analysis of Archamoebae (Amoebozoa: Conosa) robustly reveals its phylogeny and shows that Entamoebidae represents a deep lineage of the group |url=https://www.sciencedirect.com/science/article/pii/S1055790316000270 |journal=Molecular Phylogenetics and Evolution |language=en |volume=98 |pages=41–51 |doi=10.1016/j.ympev.2016.01.011 |issn=1055-7903}}</ref> It likely had mitochondria, while mitochondrial reduction has resulted in both mitochondriate and amitochondriate members today.<ref name="pmid118306644" /> The ancestral biflagellate condition is seen in some extant Conosa forms.<ref name=":13" /> The conical microtubular skeleton convergently evolved in Archamoebae and Variosea, but not in Mycetozoa.<ref name=":13" / |
The last common ancestor of Conosa was likely an aerobic [[protist]] with anterior and recurrent flagellum.<ref name=":03">{{Cite journal |last=Pánek |first=Tomáš |last2=Zadrobílková |first2=Eliška |last3=Walker |first3=Giselle |last4=Brown |first4=Matthew W. |last5=Gentekaki |first5=Eleni |last6=Hroudová |first6=Miluše |last7=Kang |first7=Seungho |last8=Roger |first8=Andrew J. |last9=Tice |first9=Alexander K. |last10=Vlček |first10=Čestmír |last11=Čepička |first11=Ivan |date=2016-05-01 |title=First multigene analysis of Archamoebae (Amoebozoa: Conosa) robustly reveals its phylogeny and shows that Entamoebidae represents a deep lineage of the group |url=https://www.sciencedirect.com/science/article/pii/S1055790316000270 |journal=Molecular Phylogenetics and Evolution |language=en |volume=98 |pages=41–51 |doi=10.1016/j.ympev.2016.01.011 |issn=1055-7903}}</ref> It likely had mitochondria, while mitochondrial reduction has resulted in both mitochondriate and amitochondriate members today.<ref name="pmid118306644" /> The ancestral biflagellate condition is seen in some extant Conosa forms.<ref name=":13" /> The conical microtubular skeleton convergently evolved in Archamoebae and Variosea, but not in Mycetozoa.<ref name=":13" /> |
||
{{clade|{{clade|label1=[[Discosea]]|1={{clade|1=[[Flabellinia]]|2=[[Centramoebia]]}}|barbegin1=grey|label2=[[Tevosa]]|2={{clade|label1=[[Tubulinea]]|bar1=grey|1={{clade|1=[[Corycidia]]|2={{clade|1=[[Echinamoebia]]|2=[[Elardia]]}}}}|label2=[[Evosea]]|2={{clade|1=[[Cutosea]]|barend1=grey|label2=[[Conosa]]|2={{clade|1=[[Archamoebea]]|label2=[[Semiconosia]]|2={{clade|1=[[Variosea]]|label2=[[Mycetozoa]]|2={{clade|1=[[Dictyostelea]]|2={{clade|1=[[Ceratiomyxea]]|2=[[Myxogastrea]]}}}}}}}}}} |
|||
}} |
|||
}}|[[Opisthokonta]]|style=font-size:90%;|label1=[[Amoebozoa]]}} |
|||
=== Gallery === |
|||
<gallery> |
|||
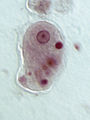
File:Entamoeba histolytica.jpg|''[[Entamoeba histolytica]]'' [[trophozoite]] |
File:Entamoeba histolytica.jpg|''[[Entamoeba histolytica]]'' [[trophozoite]] |
||
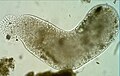
Pelomyxa palustris.jpg|''[[Pelomyxa palustris]]'' |
Pelomyxa palustris.jpg|''[[Pelomyxa palustris]]'' |
||
Revision as of 22:34, 24 April 2023
| Conosa | |
|---|---|

| |
| Dictyostelium discoideum | |
| Scientific classification | |
| Domain: | Eukaryota |
| Phylum: | Amoebozoa |
| Clade: | Evosea |
| Subphylum: | Conosa Cavalier-Smith, 1998 |
| Infraphyla | |
Conosa is a grouping of Amoebozoa. It is subdivided into three groups – Archamoeba (monophyletic), Variosea (paraphyletic) and Mycetozoa (polyphyletic).[1][2]
In some classifications, the mycetozoan Myxogastria and Dictyostelia are united in Macromycetozoa.
Conosa includes the species Dictyostelium discoideum and Entamoeba histolytica, among others.[3]
Conosa are morphologically defined by a conical microtubular structure,[4][5][6] and have been found to be monophyletic.[4][7]
Characteristics
The Conosa group was first proposed by Thomas Cavalier-Smith in 1998 as a subphylum of Amoebozoa.[8] Cavalier-Smith originally separated this group into 2 infraphyla: Archamoebae and Mycetozoa[9]. This clade is morphologically defined by their complex microtubular skeleton that forms a partial or complete cone.[10][11] They have a monolayer of microtubules that surround at least some of the anterior end of the cell and diverge into a cone shape towards the nucleus at the posterior end.[9][11] This cone of microtubles usually starts at a single centriole and extends towards the nucleus.[9] They also have a lateral microtubular ribbon towards the cell surface.[9] They can exist as aggregate aerobes with mitochondria and also as solitary anaerobes with no mitochondria or peroxisomes.[9] There are mitochondriate and amitochondriate members, as well as free living and parasitic representatives.[12]
Conosa are separated from Lobosa, the other Amoebozoa subphylum, by morphological characteristics and genomic differences. Conosa have both amoeboid and flagellate forms or stages and more pointed pseudopodia with branches. In contrast, Lobosa are entirely amoeboid with broad pseudopodia.[10] Conosa's flagella are artifacts of their ancestral conditions and are seen in trophic and swarm cell phases.[10] Flagellate Conosa have a cone-shaped microtubular skeleton, and non-ciliate forms contain extensive microtubes in the cytoplasm, both of which are not seen in Lobosa.[10]
Phylogeny
While morphological characteristics like pseudopodia and body shape, flagella, and cytoplasm properties have not been regarded as convincing taxonomic suggestions[13], emerging sequencing data is being used to support Conosa’s monophyly. A study using several hundred phylogenetic markers of 30 species found Conosa to be monophyletic as representatives of Mycetozoa, Entamoebidae, and Pelobionta grouped together using several amino acid sequencing analysis methods.[13] The monophyly of Conosa and the Archamoebea infraphyla was also supported by cDNA sequencing of 17 Amoebozoans.[14]
The last common ancestor of Conosa was likely an aerobic protist with anterior and recurrent flagellum.[15] It likely had mitochondria, while mitochondrial reduction has resulted in both mitochondriate and amitochondriate members today.[13] The ancestral biflagellate condition is seen in some extant Conosa forms.[14] The conical microtubular skeleton convergently evolved in Archamoebae and Variosea, but not in Mycetozoa.[14]
Gallery
References
- ^ Cavalier-Smith T (August 1998). "A revised six-kingdom system of life". Biol Rev Camb Philos Soc. 73 (3): 203–66. doi:10.1111/j.1469-185X.1998.tb00030.x. PMID 9809012. S2CID 6557779. Archived from the original on 2012-12-05.
- ^ Bapteste E, Brinkmann H, Lee JA, et al. (February 2002). "The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba". Proc. Natl. Acad. Sci. U.S.A. 99 (3): 1414–9. Bibcode:2002PNAS...99.1414B. doi:10.1073/pnas.032662799. PMC 122205. PMID 11830664.
- ^ Song J, Xu Q, Olsen R, et al. (December 2005). "Comparing the Dictyostelium and Entamoeba genomes reveals an ancient split in the Conosa lineage". PLOS Comput. Biol. 1 (7): e71. Bibcode:2005PLSCB...1...71S. doi:10.1371/journal.pcbi.0010071. PMC 1314882. PMID 16362072.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Cavalier-Smith, Thomas; Fiore-Donno, Anna Maria; Chao, Ema; Kudryavtsev, Alexander; Berney, Cédric; Snell, Elizabeth A.; Lewis, Rhodri (2015-02). "Multigene phylogeny resolves deep branching of Amoebozoa". Molecular Phylogenetics and Evolution. 83: 293–304. doi:10.1016/j.ympev.2014.08.011.
{{cite journal}}: Check date values in:|date=(help) - ^ Pánek, Tomáš; Zadrobílková, Eliška; Walker, Giselle; Brown, Matthew W.; Gentekaki, Eleni; Hroudová, Miluše; Kang, Seungho; Roger, Andrew J.; Tice, Alexander K.; Vlček, Čestmír; Čepička, Ivan (2016-05-01). "First multigene analysis of Archamoebae (Amoebozoa: Conosa) robustly reveals its phylogeny and shows that Entamoebidae represents a deep lineage of the group". Molecular Phylogenetics and Evolution. 98: 41–51. doi:10.1016/j.ympev.2016.01.011. ISSN 1055-7903.
- ^ Cavalier-Smith T (August 1998). "A revised six-kingdom system of life". Biol Rev Camb Philos Soc. 73 (3): 203–66. doi:10.1111/j.1469-185X.1998.tb00030.x. PMID 9809012. S2CID 6557779. Archived from the original on 2012-12-05.
- ^ Bapteste E, Brinkmann H, Lee JA, et al. (February 2002). "The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba". Proc. Natl. Acad. Sci. U.S.A. 99 (3): 1414–9. Bibcode:2002PNAS...99.1414B. doi:10.1073/pnas.032662799. PMC 122205. PMID 11830664.
- ^ Cavalier-Smith, T. (2007-01-11). "A revised six-kingdom system of life". Biological Reviews. 73 (3): 203–266. doi:10.1111/j.1469-185X.1998.tb00030.x.
- ^ a b c d e Cavalier-Smith T (August 1998). "A revised six-kingdom system of life". Biol Rev Camb Philos Soc. 73 (3): 203–66. doi:10.1111/j.1469-185X.1998.tb00030.x. PMID 9809012. S2CID 6557779. Archived from the original on 2012-12-05.
- ^ a b c d Cavalier-Smith, Thomas; Fiore-Donno, Anna Maria; Chao, Ema; Kudryavtsev, Alexander; Berney, Cédric; Snell, Elizabeth A.; Lewis, Rhodri (2015-02). "Multigene phylogeny resolves deep branching of Amoebozoa". Molecular Phylogenetics and Evolution. 83: 293–304. doi:10.1016/j.ympev.2014.08.011.
{{cite journal}}: Check date values in:|date=(help) - ^ a b Pánek, Tomáš; Zadrobílková, Eliška; Walker, Giselle; Brown, Matthew W.; Gentekaki, Eleni; Hroudová, Miluše; Kang, Seungho; Roger, Andrew J.; Tice, Alexander K.; Vlček, Čestmír; Čepička, Ivan (2016-05-01). "First multigene analysis of Archamoebae (Amoebozoa: Conosa) robustly reveals its phylogeny and shows that Entamoebidae represents a deep lineage of the group". Molecular Phylogenetics and Evolution. 98: 41–51. doi:10.1016/j.ympev.2016.01.011. ISSN 1055-7903.
- ^ Bapteste E, Brinkmann H, Lee JA, et al. (February 2002). "The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba". Proc. Natl. Acad. Sci. U.S.A. 99 (3): 1414–9. Bibcode:2002PNAS...99.1414B. doi:10.1073/pnas.032662799. PMC 122205. PMID 11830664.
- ^ a b c Bapteste E, Brinkmann H, Lee JA, et al. (February 2002). "The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba". Proc. Natl. Acad. Sci. U.S.A. 99 (3): 1414–9. Bibcode:2002PNAS...99.1414B. doi:10.1073/pnas.032662799. PMC 122205. PMID 11830664.
- ^ a b c Cavalier-Smith, Thomas; Fiore-Donno, Anna Maria; Chao, Ema; Kudryavtsev, Alexander; Berney, Cédric; Snell, Elizabeth A.; Lewis, Rhodri (2015-02). "Multigene phylogeny resolves deep branching of Amoebozoa". Molecular Phylogenetics and Evolution. 83: 293–304. doi:10.1016/j.ympev.2014.08.011.
{{cite journal}}: Check date values in:|date=(help) - ^ Pánek, Tomáš; Zadrobílková, Eliška; Walker, Giselle; Brown, Matthew W.; Gentekaki, Eleni; Hroudová, Miluše; Kang, Seungho; Roger, Andrew J.; Tice, Alexander K.; Vlček, Čestmír; Čepička, Ivan (2016-05-01). "First multigene analysis of Archamoebae (Amoebozoa: Conosa) robustly reveals its phylogeny and shows that Entamoebidae represents a deep lineage of the group". Molecular Phylogenetics and Evolution. 98: 41–51. doi:10.1016/j.ympev.2016.01.011. ISSN 1055-7903.