Butyric acid: Difference between revisions
Citation bot (talk | contribs) m Citation maintenance. [U78]Added: doi, last1, first1, last2, first2, last3, first3, last4, first4, last5, first5, last6, first6, last7, first7, last8, first8, last9, first9, pmc. Formatted: title, journal, year, volume, issue, pages, author. You can |
→Butyric acid function/activity: consistency |
||
| Line 117: | Line 117: | ||
Highly-fermentable fibers like [[resistant starch]], [[oat bran]], and [[pectin]] are transformed by [[gut flora|colonic bacteria]] into [[short chain fatty acid]]s including butyrate. One study found that resistant starch consistently produces more butyrate than other types of dietary fiber.<ref>{{cite journal | author = Cummings JH, Macfarlane GT, Englyst HN | title = Prebiotic digestion and fermentation | journal = American Journal of Clinical Nutrition | year=2001 |volume=73 | issue=suppl | pages=415S–20S}}</ref> |
Highly-fermentable fibers like [[resistant starch]], [[oat bran]], and [[pectin]] are transformed by [[gut flora|colonic bacteria]] into [[short chain fatty acid]]s including butyrate. One study found that resistant starch consistently produces more butyrate than other types of dietary fiber.<ref>{{cite journal | author = Cummings JH, Macfarlane GT, Englyst HN | title = Prebiotic digestion and fermentation | journal = American Journal of Clinical Nutrition | year=2001 |volume=73 | issue=suppl | pages=415S–20S}}</ref> |
||
The role of butyrate changes depending on its role in cancer or normal cells. This is known as the "butyrate paradox". Butyrate inhibits colonic tumor cells but promotes healthy colonic epithelial cells,<ref>{{cite journal | pmid = 19707587 | url = http://www.plosone.org/article/info:doi%2F10.1371%2Fjournal.pone.0006759 | doi = 10.1371/journal.pone.0006759 | year = 2009 | last1 = Vanhoutvin | first1 = SA | last2 = Troost | first2 = FJ | last3 = Hamer | first3 = HM | last4 = Lindsey | first4 = PJ | last5 = Koek | first5 = GH | last6 = Jonkers | first6 = DM | last7 = Kodde | first7 = A | last8 = Venema | first8 = K | last9 = Brummer | first9 = RJ | title = Butyrate-induced transcriptional changes in human colonic mucosa. | volume = 4 | issue = 8 | pages = e6759 | pmc = 2727000 | journal = PloS one}}</ref> but the signaling mechanism is not well understood.<ref>{{Cite journal | url = http://www.jbc.org/content/279/35/36680.full.pdf | title = Oncogenic Ras Promotes Butyrate-induced Apoptosis through Inhibition of Gelsolin Expression | pmid = 15213223 | year = 2004 | last1 = Klampfer | first1 = L | last2 = Huang | first2 = J | last3 = Sasazuki | first3 = T | last4 = Shirasawa | first4 = S | last5 = Augenlicht | first5 = L | volume = 279 | issue = 35 | pages = 36680–8 | doi = 10.1074/jbc.M405197200 | journal = The Journal of biological chemistry}}</ref> A review suggested that the chemopreventive benefits of butanoate depend in part on amount, time of exposure with respect to the tumorigenic process, and the type of fat in the diet.<ref>{{cite journal | url = http://jn.nutrition.org/cgi/content/full/134/2/479 |title = Microbial Degradation Products Influence Colon Cancer Risk: the Butyrate Controversy | volume = 134 | issue = 2 | pages = 479 | journal = Journal of Nutrition | pmid = 14747692 | author = Lupton, Joanne R. | year = 2004}}</ref> Low carbohydrate diets like the [[Atkins diet]] are known to reduce the amount of |
The role of butyrate changes depending on its role in cancer or normal cells. This is known as the "butyrate paradox". Butyrate inhibits colonic tumor cells but promotes healthy colonic epithelial cells,<ref>{{cite journal | pmid = 19707587 | url = http://www.plosone.org/article/info:doi%2F10.1371%2Fjournal.pone.0006759 | doi = 10.1371/journal.pone.0006759 | year = 2009 | last1 = Vanhoutvin | first1 = SA | last2 = Troost | first2 = FJ | last3 = Hamer | first3 = HM | last4 = Lindsey | first4 = PJ | last5 = Koek | first5 = GH | last6 = Jonkers | first6 = DM | last7 = Kodde | first7 = A | last8 = Venema | first8 = K | last9 = Brummer | first9 = RJ | title = Butyrate-induced transcriptional changes in human colonic mucosa. | volume = 4 | issue = 8 | pages = e6759 | pmc = 2727000 | journal = PloS one}}</ref> but the signaling mechanism is not well understood.<ref>{{Cite journal | url = http://www.jbc.org/content/279/35/36680.full.pdf | title = Oncogenic Ras Promotes Butyrate-induced Apoptosis through Inhibition of Gelsolin Expression | pmid = 15213223 | year = 2004 | last1 = Klampfer | first1 = L | last2 = Huang | first2 = J | last3 = Sasazuki | first3 = T | last4 = Shirasawa | first4 = S | last5 = Augenlicht | first5 = L | volume = 279 | issue = 35 | pages = 36680–8 | doi = 10.1074/jbc.M405197200 | journal = The Journal of biological chemistry}}</ref> A review suggested that the chemopreventive benefits of butanoate depend in part on amount, time of exposure with respect to the tumorigenic process, and the type of fat in the diet.<ref>{{cite journal | url = http://jn.nutrition.org/cgi/content/full/134/2/479 |title = Microbial Degradation Products Influence Colon Cancer Risk: the Butyrate Controversy | volume = 134 | issue = 2 | pages = 479 | journal = Journal of Nutrition | pmid = 14747692 | author = Lupton, Joanne R. | year = 2004}}</ref> Low carbohydrate diets like the [[Atkins diet]] are known to reduce the amount of butyrate produced in the colon.{{Citation needed|date=November 2009}} |
||
'''[[HDAC inhibitor]]''': Butyric acid has been associated with the ability to inhibit the function of [[histone deacetylase]] enzymes, thereby favoring an acetylated state of [[histone]]s in the cell. Acetylated histones have a lower affinity for [[DNA]] than non-acetylated histones, due to the neutralisation of [[electrostatic]] charge interactions. In general, it is thought that [[transcription factors]] will be unable to access regions where histones are tightly associated with DNA (i.e. non-acetylated, e.g., heterochromatin). Therefore, it is thought that butyric acid enhances the transcriptional activity at promoters, which are typically silenced/downregulated due to histone deacetylase activity. |
'''[[HDAC inhibitor]]''': Butyric acid has been associated with the ability to inhibit the function of [[histone deacetylase]] enzymes, thereby favoring an acetylated state of [[histone]]s in the cell. Acetylated histones have a lower affinity for [[DNA]] than non-acetylated histones, due to the neutralisation of [[electrostatic]] charge interactions. In general, it is thought that [[transcription factors]] will be unable to access regions where histones are tightly associated with DNA (i.e. non-acetylated, e.g., heterochromatin). Therefore, it is thought that butyric acid enhances the transcriptional activity at promoters, which are typically silenced/downregulated due to histone deacetylase activity. |
||
Revision as of 15:05, 6 March 2010
| |||

| |||
| Names | |||
|---|---|---|---|
| IUPAC name
Butanoic acid
| |||
| Other names
Butyric acid; 1-Propanecarboxylic acid; Propanecarboxylic acid; C4:0 (Lipid numbers)
| |||
| Identifiers | |||
3D model (JSmol)
|
|||
| ChemSpider | |||
| ECHA InfoCard | 100.003.212 | ||
| MeSH | Butyric+acid | ||
PubChem CID
|
|||
CompTox Dashboard (EPA)
|
|||
| |||
| |||
| Properties | |||
| C4H8O2 | |||
| Molar mass | 88.106 g·mol−1 | ||
| Appearance | Colorless liquid | ||
| Density | 0.9595 g/mL | ||
| Melting point | −7.9 °C (17.8 °F; 265.2 K) | ||
| Boiling point | 163.5 °C (326.3 °F; 436.6 K) | ||
| miscible | |||
| Solubility | methanol 10.94 M [1] | ||
Refractive index (nD)
|
1.3980 (19 °C) | ||
| Viscosity | 0.1529 cP | ||
| Hazards | |||
| Flash point | 72 °C | ||
| Related compounds | |||
Other anions
|
butyrate | ||
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |||
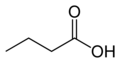
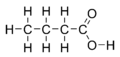
Butyric acid (from Greek βούτυρος = butter), also known under the systematic name butanoic acid, is a carboxylic acid with the structural formula CH3CH2CH2-COOH. Salts and esters of butyric acid are known as butyrates or butanoates. Butyric acid is found in butter, parmesan cheese, vomit, and as a product of anaerobic fermentation (including in the Colon (anatomy) and as body odor). It has an unpleasant smell and acrid taste, with a sweetish aftertaste (similar to ether). It can be detected by mammals with good scent detection abilities (such as dogs) at 10 ppb, whereas humans can detect it in concentrations above 10 ppm.
Chemistry
Butyric acid is a fatty acid occurring in the form of esters in animal fats and plant oils. The triglyceride of butyric acid makes up 3% to 4% of butter. When butter goes rancid, butyric acid is liberated from the glyceride by hydrolysis leading to the unpleasant odor. It is an important member of the fatty acid sub-group called short chain fatty acids. Butyric acid is a weak acid with a pKa of 4.82, similar to acetic acid which has pKa 4.76.[2] The similar strength of these acids results from their common -CH2COOH terminal structure.[3] Pure butyric acid is 10.9 molar.
The acid is an oily colorless liquid that is easily soluble in water, ethanol, and ether, and can be separated from an aqueous phase by saturation with salts such as calcium chloride. Potassium dichromate and sulfuric acid oxidize it to carbon dioxide and acetic acid, while alkaline potassium permanganate oxidizes it to carbon dioxide. The calcium salt, Ca(C4H7O2)2·H2O, is less soluble in hot water than in cold.
Butyric acid has a structural isomer called isobutyric acid (2-methylpropanoic acid).
Production
It is industrially prepared by the fermentation of sugar or starch, brought about by the addition of putrefying cheese, with calcium carbonate added to neutralize the acids formed in the process. The butyric fermentation of starch is aided by the direct addition of Bacillus subtilis. Salts and esters of the acid are called butanoates.
Butyric acid or fermentation butyric acid is also found as a hexyl ester (hexyl butanoate) in the oil of Heracleum giganteum (a type of hogweed) and as an octyl ester (octyl butanoate) in parsnip (Pastinaca sativa); it has also been noticed in the fluors of the flesh and in perspiration.
Uses
Butyric acid is used in the preparation of various butanoate esters. Low-molecular-weight esters of butyric acid, such as methyl butanoate, have mostly pleasant aromas or tastes. As a consequence, they find use as food and perfume additives.
Due to the powerful odor it has also been used as a successful fishing bait additive[4] and by anti-whaling protestors against Japanese whaling ships.[5]
Biological functionality
Butanoate fermentation
Butanoate is produced as end-product of a fermentation process solely performed by obligate anaerobic bacteria. Fermented Kombucha "tea" includes butyric acid as a result of the fermentation. This fermentation pathway was discovered by Louis Pasteur in 1861. Examples of butanoate-producing species of bacteria:
- Clostridium acetobutylicum
- Clostridium butyricum
- Clostridium kluyveri
- Clostridium pasteurianum
- Fusobacterium nucleatum
- Butyrivibrio fibrisolvens
- Eubacterium limosum
The pathway starts with the glycolytic cleavage of glucose to two molecules of pyruvate, as happens in most organisms. Pyruvate is then oxidized into acetyl coenzyme A using a unique mechanism that involves an enzyme system called pyruvate-ferredoxin oxidoreductase. Two molecules of carbon dioxide (CO2) and two molecules of elemental hydrogen (H2) are formed as wastes products from the cell. Then,
| Action | Responsible enzyme |
|---|---|
| Acetyl coenzyme A converts into acetoacetyl coenzyme A | acetyl-CoA-acetyl transferase |
| Acetoacetyl coenzyme A converts into β-hydroxybutyryl CoA | β-hydroxybutyryl-CoA dehydrogenase |
| β-hydroxybutyryl CoA converts into crotonyl CoA | crotonase |
| Crotonyl CoA converts into butyryl CoA (CH3CH2CH2C=O-CoA) | butyryl CoA dehydrogenase |
| A phosphate group replaces CoA to form butyryl phosphate | phosphobutyrylase |
| The phosphate group joins ADP to form ATP and butyrate | butyrate kinase |
ATP is produced, as can be seen, in the last step of the fermentation. Three molecules of ATP are produced for each glucose molecule, a relatively high yield. The balanced equation for this fermentation is
- C6H12O6 → C4H8O2 + 2CO2 + 2H2.
Acetone and butanol fermentation
Several species form acetone and butanol in an alternative pathway, which starts as butyrate fermentation. Some of these species are
- Clostridium acetobutylicum, the most prominent acetone and butanol producer, used also in industry,
- Clostridium beijerinckii,
- Clostridium tetanomorphum,
- Clostridium aurantibutyricum.
These bacteria begin with butanoate fermentation as described above, but, when the pH drops below 5, they switch into butanol and acetone production in order to prevent further lowering of the pH. Two molecules of butanol are formed for each molecule of acetone.
The change in the pathway occurs after acetoacetyl CoA formation. This intermediate then takes two possible pathways:
- acetoacetyl CoA → acetoacetate → acetone, or
- acetoacetyl CoA → butyryl CoA → butanal → butanol.
Butyric acid function/activity
Highly-fermentable fibers like resistant starch, oat bran, and pectin are transformed by colonic bacteria into short chain fatty acids including butyrate. One study found that resistant starch consistently produces more butyrate than other types of dietary fiber.[6]
The role of butyrate changes depending on its role in cancer or normal cells. This is known as the "butyrate paradox". Butyrate inhibits colonic tumor cells but promotes healthy colonic epithelial cells,[7] but the signaling mechanism is not well understood.[8] A review suggested that the chemopreventive benefits of butanoate depend in part on amount, time of exposure with respect to the tumorigenic process, and the type of fat in the diet.[9] Low carbohydrate diets like the Atkins diet are known to reduce the amount of butyrate produced in the colon.[citation needed]
HDAC inhibitor: Butyric acid has been associated with the ability to inhibit the function of histone deacetylase enzymes, thereby favoring an acetylated state of histones in the cell. Acetylated histones have a lower affinity for DNA than non-acetylated histones, due to the neutralisation of electrostatic charge interactions. In general, it is thought that transcription factors will be unable to access regions where histones are tightly associated with DNA (i.e. non-acetylated, e.g., heterochromatin). Therefore, it is thought that butyric acid enhances the transcriptional activity at promoters, which are typically silenced/downregulated due to histone deacetylase activity.
Two HDAC inhibitors, sodium butyrate (NaB) and Trichostatin A (TSA), increase lifespan in experimental animals.[10]
See also
References
This article incorporates text from a publication now in the public domain: Chisholm, Hugh, ed. (1911). Encyclopædia Britannica (11th ed.). Cambridge University Press. {{cite encyclopedia}}: Missing or empty |title= (help)
- ^ Solubility of butyric acid in methanol
- ^ "Adimix Sodium Butanoate information" (PDF).
- ^ "Using the pKa table".
- ^ Freezer Baits, nutrabaits.net
- ^ Japanese Whalers Injured by Acid-Firing Activists, newser.com, February 10, 2010
- ^ Cummings JH, Macfarlane GT, Englyst HN (2001). "Prebiotic digestion and fermentation". American Journal of Clinical Nutrition. 73 (suppl): 415S–20S.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Vanhoutvin, SA; Troost, FJ; Hamer, HM; Lindsey, PJ; Koek, GH; Jonkers, DM; Kodde, A; Venema, K; Brummer, RJ (2009). "Butyrate-induced transcriptional changes in human colonic mucosa". PloS one. 4 (8): e6759. doi:10.1371/journal.pone.0006759. PMC 2727000. PMID 19707587.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Klampfer, L; Huang, J; Sasazuki, T; Shirasawa, S; Augenlicht, L (2004). "Oncogenic Ras Promotes Butyrate-induced Apoptosis through Inhibition of Gelsolin Expression" (PDF). The Journal of biological chemistry. 279 (35): 36680–8. doi:10.1074/jbc.M405197200. PMID 15213223.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Lupton, Joanne R. (2004). "Microbial Degradation Products Influence Colon Cancer Risk: the Butyrate Controversy". Journal of Nutrition. 134 (2): 479. PMID 14747692.
- ^ Zhang, M; Poplawski, M; Yen, K; Cheng, H; Bloss, E; Zhu, X; Patel, H; Mobbs, CV; Dillin, Andy (2009). "Role of CBP and SATB-1 in aging, dietary restriction, and insulin-like signaling". PLoS biology. 7 (11): e1000245. doi:10.1371/journal.pbio.1000245. PMC 2774267. PMID 19924292.
{{cite journal}}: CS1 maint: unflagged free DOI (link)