Heritability: Difference between revisions
awkward jargon does not help anybody |
|||
| Line 12: | Line 12: | ||
[[Image:Heritability-from-twin-correlations1.jpg|300px|thumbnail|Fig 1. Heritability for nine psychological traits as estimated from twin studies. All sources are twins raised together (sample size shown inside bars). MZ: Monozygotic twin, DZ: Dizygotic twin]]In non-human populations it is often possible to collect information in a controlled way. For example among farm animals it is easy to arrange for (say) a bull sire to produce offspring from a large number of cows. Due to ethical concerns, such a degree of experimental control is impossible when gathering human data. As a result, studies of human heritability often contrast identical twins who have been separated early in life and raised in different environments (see for example Fig. 1). Such individuals have identical genotypes and can be used to separate the effects of genotype and environment. But twin analysis entails problems of its own, not the least of which is that independently raised twins shared a common prenatal environment and are also a rare commodity. |
[[Image:Heritability-from-twin-correlations1.jpg|300px|thumbnail|Fig 1. Heritability for nine psychological traits as estimated from twin studies. All sources are twins raised together (sample size shown inside bars). MZ: Monozygotic twin, DZ: Dizygotic twin]]In non-human populations it is often possible to collect information in a controlled way. For example among farm animals it is easy to arrange for (say) a bull sire to produce offspring from a large number of cows. Due to ethical concerns, such a degree of experimental control is impossible when gathering human data. As a result, studies of human heritability often contrast identical twins who have been separated early in life and raised in different environments (see for example Fig. 1). Such individuals have identical genotypes and can be used to separate the effects of genotype and environment. But twin analysis entails problems of its own, not the least of which is that independently raised twins shared a common prenatal environment and are also a rare commodity. |
||
Heritability estimates are always relative to the genetic and environmental |
Heritability estimates are always relative to the genetic and environmental factors that shaped the phenotypic variance of the samples used to make the estimates, and therefore are not absolute measurements of the contribution of genetic and environmental factors to a phenotype. Since estimates of heritability reflect the extent to which genotypic effects affect the phenotypic variance relative to the extent to which environmental effects do it, heritability estimates can be made larger by diversifying the genetic background, e.g., by using only very outbred individuals (which increases the Variance(G)) and/or by minimizing environmental effects (which decreases the Variance(E)). Smaller estimates, on the other hand, can be generated by using [[Inbreeding|inbred]] individuals (which decreases the Variance(G)) or individuals reared in very diverse environments (which increases the Variance(E)). Due to such "bckground" effects, different populations of a species might have different heritabilities even for the same trait. |
||
Because of the contextual nature of measured heritabilities, paradoxes often arise. For example, the heritability of a trait could be near 100% in one study and close to zero in another. In one study, e.g., a group of unrelated army recruits may be given identical training and nutrition and then their muscular strength may be measured. The variation in strength observed after the (identical) training will translate into a high heritability estimate. In another study, whose purpose might be to assess the efficacy of various workout regimes or nutritional programs, study subjects may be first chosen to match each other as closely as possible in prior physical characteristics before some of them are put onto Program A and others onto Program B, and this will lead to a low heritability estimate. |
Because of the contextual nature of measured heritabilities, paradoxes often arise. For example, the heritability of a trait could be near 100% in one study and close to zero in another. In one study, e.g., a group of unrelated army recruits may be given identical training and nutrition and then their muscular strength may be measured. The variation in strength observed after the (identical) training will translate into a high heritability estimate. In another study, whose purpose might be to assess the efficacy of various workout regimes or nutritional programs, study subjects may be first chosen to match each other as closely as possible in prior physical characteristics before some of them are put onto Program A and others onto Program B, and this will lead to a low heritability estimate. |
||
Revision as of 05:21, 24 April 2006
In genetics, heritability is the proportion of phenotypic variation in a population that is due to genetic variation. Variation among individuals may be due to genetic and/or environmental factors. Heritability analyses estimate the relative importance of variation in each of these factors.
The equation for heritability is derived from the equation Phenotype (P) = Genotype (G) + Environment (E):
- H2 = Variance(G) / Variance(P)
This is called the "broad-sense" heritability and reflects all possible genetic contributions to the population's phenotypic variance. Included are effects due to additively acting variation, to variants that are dominant or act epistatically in combination with others, as well as maternal and paternal effects that are not necessarily passed to one's progeny. The "narrow-sense" heritability (h2), quantifies only the portion of the phenotypic variation] that is of additive nature (note upper case H2 for broad sense, lower case h2 for narrow sense). When one is interested, e.g., in improving livestock via artificial selection, knowing the narrow-sense heritability of the trait of interest will allow one to predict by how much the mean of the trait will increase in the next generation as a function of how much the mean of the selected parents differs from the mean of the population from which the selected parents were chosen. Indeed, from the observed "response to selection" one can estimate the realized heritability, which is itself an estimate of the narrow-sense heritability.
Estimating heritability
Estimating heritability is not a simple process, however, since only P can be observed or measured directly. Measuring the genetic and environmental variance requires sophisticated statistical methods one which being technique called "variance-component estimation". This methodology delivers better estimates with large volumes of data, as do many statistical techniques, and it is specially powerful when it is fed data from closely related individuals - such as brothers, sisters, parents and children, rather than from distantly related ones.

In non-human populations it is often possible to collect information in a controlled way. For example among farm animals it is easy to arrange for (say) a bull sire to produce offspring from a large number of cows. Due to ethical concerns, such a degree of experimental control is impossible when gathering human data. As a result, studies of human heritability often contrast identical twins who have been separated early in life and raised in different environments (see for example Fig. 1). Such individuals have identical genotypes and can be used to separate the effects of genotype and environment. But twin analysis entails problems of its own, not the least of which is that independently raised twins shared a common prenatal environment and are also a rare commodity.
Heritability estimates are always relative to the genetic and environmental factors that shaped the phenotypic variance of the samples used to make the estimates, and therefore are not absolute measurements of the contribution of genetic and environmental factors to a phenotype. Since estimates of heritability reflect the extent to which genotypic effects affect the phenotypic variance relative to the extent to which environmental effects do it, heritability estimates can be made larger by diversifying the genetic background, e.g., by using only very outbred individuals (which increases the Variance(G)) and/or by minimizing environmental effects (which decreases the Variance(E)). Smaller estimates, on the other hand, can be generated by using inbred individuals (which decreases the Variance(G)) or individuals reared in very diverse environments (which increases the Variance(E)). Due to such "bckground" effects, different populations of a species might have different heritabilities even for the same trait.
Because of the contextual nature of measured heritabilities, paradoxes often arise. For example, the heritability of a trait could be near 100% in one study and close to zero in another. In one study, e.g., a group of unrelated army recruits may be given identical training and nutrition and then their muscular strength may be measured. The variation in strength observed after the (identical) training will translate into a high heritability estimate. In another study, whose purpose might be to assess the efficacy of various workout regimes or nutritional programs, study subjects may be first chosen to match each other as closely as possible in prior physical characteristics before some of them are put onto Program A and others onto Program B, and this will lead to a low heritability estimate.
In the case of scholastic ability, how well one does in the final school exams depends on both what and how well one was taught, how hard one has studied, how ‘naturally’ smart one is and, of course, on a fair bit on luck. The actual heritability estimate will depend on the subjects used (reflecting genetic variation) and the testing conditions (reflecting environmental variation).
Much the same goes for intelligence tests. The conclusions from studies involving intelligence tests often conclude that intelligence has high heritability. This is probably due to inherent problems with human twin studies, as well as reflecting a high level of genetic variation for many human traits, and corresponding lower environmental variation within the confines of the test.
In human genetics, much use is made of twin studies in the analysis of heritability – monozygous (‘identical’) twins are clones of each other, and have effectively identical genotypes, and similarities between sets of monozygous twins can be compared with those between dizygous (‘fraternal’, or non-identical) twins, who have only a ½ coefficient of relatedness to each other. Using twins also introduces certain unique challenges, such as a common prenatal environment and intrauterine competition.
Heritability is often misunderstood when presented in the non-scientific media. Heritability only describes how much variation in the phenotype is attributable to variation in genotypes compared to the variation in environments. Heritability does not quantify the extent to which genes and environment actually determine a phenotype, let alone the extent to which changes in genes and environment could change phenotypic values (see Reaction norm).
Estimation methods
There are essentially two schools of thought regarding estimation of heritability.
One school of thought was developed by Sewall Wright at The University of Chicago, and further popularized by C. C. Li (University of Chicago) and J. L. Lush (Iowa State University). It is based on the analysis of correlations and, by extension, regression. Path Analysis was developed by Sewall Wright as a way of estimating heritability.
The second was originally developed by R. A. Fisher and expanded at The University of Edinburgh, Iowa State University, and North Carolina State University, as well as other schools. It is based on the analysis of variance of breeding studies, using the intraclass correlation of relatives. Various methods of estimating components of variance (and, hence, heritability) from ANOVA are used in these analyses.
Regression/correlation methods of estimation
The first school of estimation uses regression and correlation to estimate heritability.
Selection experiments
Calculating the strength of selection, S (the difference in mean trait between the population as a whole and the selected parents of the next generation, also called the selection differential [1]) and response to selection R (the difference in offspring and whole parental generation mean trait) in an artificial selection experiment will allow calculation of realized heritability as the response to selection relative to the strength of selection, h2=R/S as in Fig. 2.
Comparison of close relatives
In the comparison of relatives, we find that in general,
where r can be thought of as the coefficient of relatedness, b is the coefficient of regression and t the coefficient of correlation.
Parent-offspring regression
Heritability may be estimated by comparing parent and offspring traits (as In Fig 3). The slope of the line (0.57) approximates the heritability of the trait when offspring values are regressed against the average trait in the parents. If only one parents value is used then heritability is twice the slope. (note that this is the source of the term "regression", since the offspring values always tend to regress to the mean value for the population,ie the slope is always less than one).
Full-sib comparison
Full-sib designs compare phenotypic traits of siblings that share a mother and a father with other sibling groups.
This section needs expansion. You can help by adding to it. |
Half-sib comparison
Half-sib designs compare phenotypic traits of siblings that share one parent with other sibling groups.
This section needs expansion. You can help by adding to it. |
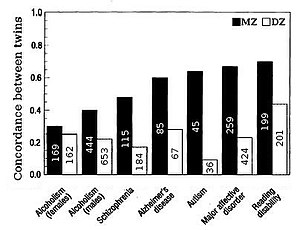
Heritability for traits in humans is most frequently estimated by comparing resemblances between twins (Fig. 1 & 4). Identical twins (MZ twins) are twice as genetically similar as fraternal twins (DZ twins) and so heritability is approximately twice the difference in correlation between MZ and DZ twins, h2=2(r(MZ)-r(DZ)). The effect of shared environment, c2, contributes to similarity between siblings due to the commonality of the environment they are raised in. Shared environment is approximated by the DZ correlation minus half heritability, which is the degee to which DZ twins share the same genes, c2=DZ-1/2h2. Unique environmental variance, e2, reflects the degree to which identical twins raised together are dissimilar, e2=1-r(MZ).
Large, complex pedigrees
This section needs expansion. You can help by adding to it. |
Analysis of variance methods of estimation
The second set of methods of estimation of heritability involves ANOVA and estimation of variance components.
Basic model
We use the basic discussion of Kempthorne (1957 [1969]). Considering only the most basic of genetic models, we can look at the quantitative contribution of a single locus with genotype Gi as
where
is the effect of genotype Gi
and is the environmental effect.
Consider an experiment with a group of sires and their progeny from random dams. Since the progeny get half of their genes from the father and half from their (random) mother, the progeny equation is
Intraclass correlations
Consider the experiment above. We have two groups of progeny we can compare. The first is comparing the various progeny for an individual sire (called within sire group). The variance will include terms for genetic variance (since they did not all get the same genotype) and environmental variance. This is thought of as an error term.
The second group of progeny are comparisons of means of half sibs with each other (called among sire group). In addition to the error term as in the within sire groups, we have an addition term due to the differences among different means of half sibs. The intraclass correlation is
The ANOVA
In an experiment with sires and progeny per sire, we can calculate the following ANOVA, using as the genetic variance and as the environmental variance:
| Source | d.f. | Mean Square | Expected Mean Square |
|---|---|---|---|
| Among sire groups | |||
| Within sire groups |
The term is the intraclass correlation among half sibs. We can easily calculate . The Expected Mean Square is calculated from the relationship of the individuals (progeny within a sire are all half-sibs, for example), and an understanding of intraclass correlations.
Model with additive and dominance terms
For a model with additive and dominance terms, but not others, the equation for a single locus is
where
is the additive effect of the ith allele, is the additive effect of the jth allele, is the dominance deviation for the ijth genotype, and is the environment.
Experiments can be run with a similar setup to the one given in Table 1. Using different relationship groups, we can evaluate different intraclass correlations. Using as the additive genetic variance and as the dominance deviation variance, intraclass correlations become linear functions of these parameters. In general,
- Intraclass correlation
where and are found as
P[alleles drawn at random from the relationship pair are identical by descent], and
P[genotypes drawn at random from the relationship pair are identical by descent].
Some common relationships and their coefficients are given in Table 2.
| Relationship | ||
|---|---|---|
| Identical Twins | ||
| Parent-Offspring | ||
| Half Siblings | ||
| Full Siblings | ||
| First Cousins | ||
| Double First Cousins |
Larger models
When a large, complex pedigree is available for estimating heritability, the most efficient use of the data is in a restricted maximum likelihood (REML) model. The raw data will usually have three or more datapoints for each individual: a code for the sire, a code for the dam and one or several trait values. Different trait values may be for different traits or for different timepoints of measurement. The currently popular methodology relies on high degrees of certainty over the identities of the sire and dam; it is not common to treat the sire identity probabilistically. This is not usually a problem, since the methodology is rarely applied to wild populations (although it has been used for several wild ungulate and bird populations), and sires are invariably known with a very high degree of certainty in artificial breeding programmes.
The pedigrees can be viewed using programs such as Pedigree Viewer [1], and analysed with programs such as ASReml or VCE [2].
External links
- Stanford Encyclopedia of Philosophy entry on Heredity and Heritability
- Quantitative Genetics Resources website, including the two volume book by Lynch and Walsh. Free access
References
- Falconer, D. S. & Mackay TFC (1996). Introduction to Quantitative Genetics. Fourth edition. Addison Wesley Longman, Harlow, Essex, U.K.
- Gillespie, G. H. (1997). Population Genetics: A Concise Guide. Johns Hopkins University Press.
- Kempthorne, O (1957 [1969]) An Introduction to Genetic Statistics. John Wiley. Reprinted, 1969 by Iowa State University Press.
- Lynch, M. & Walsh, B. 1997. Genetics and Analysis of Quantitative Traits. Sinauer Associates. ISBN 0878934812.
- Malécot, G. 1948. Les Mathématiques de l'Hérédité. Masson, Paris.
- Wahlsten, D. (1994) The intelligence of heritability. Canadian Psychology 35, 244-258.
Notes
- ^ Kempthorne (1957), page 507; or Falconer (1960[1970]), page 191, for example.


































