Adenosine deaminase: Difference between revisions
→Further reading: 10.1128/mcb.5.4.762 |
Citation bot (talk | contribs) Alter: pmid. Add: isbn, s2cid. Removed parameters. | You can use this bot yourself. Report bugs here. | Suggested by Abductive | Category:Genes on human chromosome 20 | via #UCB_Category |
||
| Line 82: | Line 82: | ||
== Mechanism of catalysis == |
== Mechanism of catalysis == |
||
The proposed mechanism for ADA-catalyzed deamination is stereospecific addition-elimination via tetrahedral intermediate.<ref name = "twentythree">{{cite journal | vauthors = Losey HC, Ruthenburg AJ, Verdine GL | title = Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA | journal = Nature Structural & Molecular Biology | volume = 13 | issue = 2 | pages = 153–159 | date = Jan 2006 | pmid = | pmc = | doi = 10.1038/nsmb1047 }}</ref> By either mechanism, Zn<sup>2+</sup> as a strong electrophile activates a water molecule, which is deprotonated by the basic Asp295 to form the attacking hydroxide.<ref name = "four"/> His238 orients the water molecule and stabilizes the charge of the attacking hydroxide. Glu217 is protonated to donate a proton to N1 of the substrate. |
The proposed mechanism for ADA-catalyzed deamination is stereospecific addition-elimination via tetrahedral intermediate.<ref name = "twentythree">{{cite journal | vauthors = Losey HC, Ruthenburg AJ, Verdine GL | title = Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA | journal = Nature Structural & Molecular Biology | volume = 13 | issue = 2 | pages = 153–159 | date = Jan 2006 | pmid = 16415880| pmc = | doi = 10.1038/nsmb1047 | s2cid = 34848284 }}</ref> By either mechanism, Zn<sup>2+</sup> as a strong electrophile activates a water molecule, which is deprotonated by the basic Asp295 to form the attacking hydroxide.<ref name = "four"/> His238 orients the water molecule and stabilizes the charge of the attacking hydroxide. Glu217 is protonated to donate a proton to N1 of the substrate. |
||
The reaction is [[stereospecificity|stereospecific]] due to the location of the zinc, Asp295, and His238 residues, which all face the B-side of the purine ring of the substrate.<ref name = "four"/> |
The reaction is [[stereospecificity|stereospecific]] due to the location of the zinc, Asp295, and His238 residues, which all face the B-side of the purine ring of the substrate.<ref name = "four"/> |
||
| Line 96: | Line 96: | ||
== Pathology == |
== Pathology == |
||
Some [[mutation]]s in the gene for adenosine deaminase cause it not to be expressed. The resulting deficiency is one cause of [[severe combined immunodeficiency]] (SCID), particularly of autosomal recessive inheritance.<ref name="pmid17181544">{{cite journal | vauthors = Sanchez JJ, Monaghan G, Børsting C, Norbury G, Morling N, Gaspar HB | title = Carrier frequency of a nonsense mutation in the adenosine deaminase (ADA) gene implies a high incidence of ADA-deficient severe combined immunodeficiency (SCID) in Somalia and a single, common haplotype indicates common ancestry | journal = Annals of Human Genetics | volume = 71 | issue = Pt 3 | pages = 336–47 | date = May 2007 | pmid = 17181544 | doi = 10.1111/j.1469-1809.2006.00338.x }}</ref> Deficient levels of ADA have also been associated with pulmonary inflammation, thymic cell death, and defective T-cell receptor signaling.<ref name = "pulmon">{{cite journal | vauthors = Blackburn MR, Kellems RE | title = Adenosine deaminase deficiency: metabolic basis of immune deficiency and pulmonary inflammation |
Some [[mutation]]s in the gene for adenosine deaminase cause it not to be expressed. The resulting deficiency is one cause of [[severe combined immunodeficiency]] (SCID), particularly of autosomal recessive inheritance.<ref name="pmid17181544">{{cite journal | vauthors = Sanchez JJ, Monaghan G, Børsting C, Norbury G, Morling N, Gaspar HB | title = Carrier frequency of a nonsense mutation in the adenosine deaminase (ADA) gene implies a high incidence of ADA-deficient severe combined immunodeficiency (SCID) in Somalia and a single, common haplotype indicates common ancestry | journal = Annals of Human Genetics | volume = 71 | issue = Pt 3 | pages = 336–47 | date = May 2007 | pmid = 17181544 | doi = 10.1111/j.1469-1809.2006.00338.x | s2cid = 34850391 }}</ref> Deficient levels of ADA have also been associated with pulmonary inflammation, thymic cell death, and defective T-cell receptor signaling.<ref name = "pulmon">{{cite journal | vauthors = Blackburn MR, Kellems RE | title = Adenosine deaminase deficiency: metabolic basis of immune deficiency and pulmonary inflammation | volume = 86 | pages = 1–41 | year = 2005 | pmid = 15705418 | pmc = | doi = 10.1016/S0065-2776(04)86001-2 | isbn = 9780120044863 | series = Advances in Immunology }}</ref><ref name = "thym">{{cite journal | vauthors = Apasov SG, Blackburn MR, Kellems RE, Smith PT, Sitkovsky MV | title = Adenosine deaminase deficiency increases thymic apoptosis and causes defective T cell receptor signaling | journal = The Journal of Clinical Investigation | volume = 108 | issue = 1 | pages = 131–141 | date = Jul 2001 | pmid = 11435465 | pmc = 209335 | doi = 10.1172/JCI10360 }}</ref> |
||
Conversely, mutations causing this enzyme to be overexpressed are one cause of [[hemolytic anemia]].<ref name="pmid3029177">{{cite journal | vauthors = Chottiner EG, Cloft HJ, Tartaglia AP, Mitchell BS | title = Elevated adenosine deaminase activity and hereditary hemolytic anemia. Evidence for abnormal translational control of protein synthesis | journal = The Journal of Clinical Investigation | volume = 79 | issue = 3 | pages = 1001–5 | date = Mar 1987 | pmid = 3029177 | pmc = 424261 | doi = 10.1172/JCI112866 }}</ref> |
Conversely, mutations causing this enzyme to be overexpressed are one cause of [[hemolytic anemia]].<ref name="pmid3029177">{{cite journal | vauthors = Chottiner EG, Cloft HJ, Tartaglia AP, Mitchell BS | title = Elevated adenosine deaminase activity and hereditary hemolytic anemia. Evidence for abnormal translational control of protein synthesis | journal = The Journal of Clinical Investigation | volume = 79 | issue = 3 | pages = 1001–5 | date = Mar 1987 | pmid = 3029177 | pmc = 424261 | doi = 10.1172/JCI112866 }}</ref> |
||
| Line 117: | Line 117: | ||
Total plasma ADA can be measured using [[high performance liquid chromatography]] or enzymatic or colorimetric techniques. Perhaps the simplest system is the measurement of the [[ammonia]] released from adenosine when broken down to inosine. After incubation of plasma with a buffered solution of adenosine the ammonia is reacted with a [[Berthelot reagent]] to form a blue colour which is proportionate to the amount of enzyme activity. To measure ADA2, [[EHNA|erythro-9-(2-hydroxy-3-nonyl) adenine]] (EHNA) is added prior to incubation so as to inhibit the enzymatic activity of ADA1.<ref name="pmid24216" /> It is the absence of ADA1 that causes [[Severe combined immunodeficiency|SCID]]. |
Total plasma ADA can be measured using [[high performance liquid chromatography]] or enzymatic or colorimetric techniques. Perhaps the simplest system is the measurement of the [[ammonia]] released from adenosine when broken down to inosine. After incubation of plasma with a buffered solution of adenosine the ammonia is reacted with a [[Berthelot reagent]] to form a blue colour which is proportionate to the amount of enzyme activity. To measure ADA2, [[EHNA|erythro-9-(2-hydroxy-3-nonyl) adenine]] (EHNA) is added prior to incubation so as to inhibit the enzymatic activity of ADA1.<ref name="pmid24216" /> It is the absence of ADA1 that causes [[Severe combined immunodeficiency|SCID]]. |
||
ADA can also be used in the workup of lymphocytic [[pleural effusion]]s or [[Ascites|peritoneal ascites]], in that such specimens with low ADA levels essentially excludes tuberculosis from consideration.<ref name="pmid12608433">{{cite journal | vauthors = Jiménez Castro D, Díaz Nuevo G, Pérez-Rodríguez E, Light RW | title = Diagnostic value of adenosine deaminase in nontuberculous lymphocytic pleural effusions | journal = Eur. Respir. J. | volume = 21 | issue = 2 | pages = 220–4 | year = 2003 | pmid = 12608433 | doi = 10.1183/09031936.03.00051603 | url = http://erj.ersjournals.com/content/21/2/220.full.pdf }}</ref> |
ADA can also be used in the workup of lymphocytic [[pleural effusion]]s or [[Ascites|peritoneal ascites]], in that such specimens with low ADA levels essentially excludes tuberculosis from consideration.<ref name="pmid12608433">{{cite journal | vauthors = Jiménez Castro D, Díaz Nuevo G, Pérez-Rodríguez E, Light RW | title = Diagnostic value of adenosine deaminase in nontuberculous lymphocytic pleural effusions | journal = Eur. Respir. J. | volume = 21 | issue = 2 | pages = 220–4 | year = 2003 | pmid = 12608433 | doi = 10.1183/09031936.03.00051603 | s2cid = 10703687 | url = http://erj.ersjournals.com/content/21/2/220.full.pdf }}</ref> |
||
[[Tuberculosis]] pleural effusions can now be diagnosed accurately by increased levels of pleural fluid adenosine deaminase, above 40 U per liter.<ref>{{cite book | first1 = F. | last1 = Brunicardi | first2 = Dana | last2 = Andersen | first3 = Timothy | last3 = Billiar | first4 = David | last4 = Dunn | first5 = John | last5 = Hunter | first6 = Raphael E. | last6 = Pollock | title = Schwartz's principles of surgery | date = 2005 | publisher = McGraw-Hill Professional | location = New York | isbn = 978-0071410908 | edition = 8th | chapter = Chapter 18, question 16 | name-list-format = vanc }}</ref> |
[[Tuberculosis]] pleural effusions can now be diagnosed accurately by increased levels of pleural fluid adenosine deaminase, above 40 U per liter.<ref>{{cite book | first1 = F. | last1 = Brunicardi | first2 = Dana | last2 = Andersen | first3 = Timothy | last3 = Billiar | first4 = David | last4 = Dunn | first5 = John | last5 = Hunter | first6 = Raphael E. | last6 = Pollock | title = Schwartz's principles of surgery | date = 2005 | publisher = McGraw-Hill Professional | location = New York | isbn = 978-0071410908 | edition = 8th | chapter = Chapter 18, question 16 | name-list-format = vanc }}</ref> |
||
| Line 132: | Line 132: | ||
{{refbegin|33em}} |
{{refbegin|33em}} |
||
* {{cite journal | vauthors = da Cunha JG | title = [Adenosine deaminase. A pluridisciplinary enzyme] | journal = Acta Médica Portuguesa | volume = 4 | issue = 6 | pages = 315–23 | year = 1992 | pmid = 1807098 | doi = }} |
* {{cite journal | vauthors = da Cunha JG | title = [Adenosine deaminase. A pluridisciplinary enzyme] | journal = Acta Médica Portuguesa | volume = 4 | issue = 6 | pages = 315–23 | year = 1992 | pmid = 1807098 | doi = }} |
||
* {{cite journal | vauthors = Franco R, Casadó V, Ciruela F, Saura C, Mallol J, Canela EI, Lluis C | title = Cell surface adenosine deaminase: much more than an ectoenzyme | journal = Progress in Neurobiology | volume = 52 | issue = 4 | pages = 283–94 | date = Jul 1997 | pmid = 9247966 | doi = 10.1016/S0301-0082(97)00013-0 }} |
* {{cite journal | vauthors = Franco R, Casadó V, Ciruela F, Saura C, Mallol J, Canela EI, Lluis C | title = Cell surface adenosine deaminase: much more than an ectoenzyme | journal = Progress in Neurobiology | volume = 52 | issue = 4 | pages = 283–94 | date = Jul 1997 | pmid = 9247966 | doi = 10.1016/S0301-0082(97)00013-0 | s2cid = 40318396 }} |
||
* {{cite journal | vauthors = Valenzuela A, Blanco J, Callebaut C, Jacotot E, Lluis C, Hovanessian AG, Franco R | title = HIV-1 envelope gp120 and viral particles block adenosine deaminase binding to human CD26 | journal = Advances in Experimental Medicine and Biology | volume = 421 | issue = | pages = 185–92 | year = 1997 | pmid = 9330696 | doi = 10.1007/978-1-4757-9613-1_24 }} |
* {{cite journal | vauthors = Valenzuela A, Blanco J, Callebaut C, Jacotot E, Lluis C, Hovanessian AG, Franco R | title = HIV-1 envelope gp120 and viral particles block adenosine deaminase binding to human CD26 | journal = Advances in Experimental Medicine and Biology | volume = 421 | issue = | pages = 185–92 | year = 1997 | pmid = 9330696 | doi = 10.1007/978-1-4757-9613-1_24 | isbn = 978-1-4757-9615-5 }} |
||
* {{cite journal | vauthors = Moriwaki Y, Yamamoto T, Higashino K | title = Enzymes involved in purine metabolism--a review of histochemical localization and functional implications | journal = Histology and Histopathology | volume = 14 | issue = 4 | pages = 1321–40 | date = Oct 1999 | pmid = 10506947 | doi = }} |
* {{cite journal | vauthors = Moriwaki Y, Yamamoto T, Higashino K | title = Enzymes involved in purine metabolism--a review of histochemical localization and functional implications | journal = Histology and Histopathology | volume = 14 | issue = 4 | pages = 1321–40 | date = Oct 1999 | pmid = 10506947 | doi = }} |
||
* {{cite journal | vauthors = Hirschhorn R | title = Identification of two new missense mutations (R156C and S291L) in two ADA- SCID patients unusual for response to therapy with partial exchange transfusions | journal = Human Mutation | volume = 1 | issue = 2 | pages = 166–8 | year = 1993 | pmid = 1284479 | doi = 10.1002/humu.1380010214 }} |
* {{cite journal | vauthors = Hirschhorn R | title = Identification of two new missense mutations (R156C and S291L) in two ADA- SCID patients unusual for response to therapy with partial exchange transfusions | journal = Human Mutation | volume = 1 | issue = 2 | pages = 166–8 | year = 1993 | pmid = 1284479 | doi = 10.1002/humu.1380010214 | s2cid = 44617309 }} |
||
* {{cite journal | vauthors = Berkvens TM, van Ormondt H, Gerritsen EJ, Khan PM, van der Eb AJ | title = Identical 3250-bp deletion between two AluI repeats in the ADA genes of unrelated ADA-SCID patients | journal = Genomics | volume = 7 | issue = 4 | pages = 486–90 | date = Aug 1990 | pmid = 1696926 | doi = 10.1016/0888-7543(90)90190-6 }} |
* {{cite journal | vauthors = Berkvens TM, van Ormondt H, Gerritsen EJ, Khan PM, van der Eb AJ | title = Identical 3250-bp deletion between two AluI repeats in the ADA genes of unrelated ADA-SCID patients | journal = Genomics | volume = 7 | issue = 4 | pages = 486–90 | date = Aug 1990 | pmid = 1696926 | doi = 10.1016/0888-7543(90)90190-6 }} |
||
* {{cite journal | vauthors = Aran JM, Colomer D, Matutes E, Vives-Corrons JL, Franco R | title = Presence of adenosine deaminase on the surface of mononuclear blood cells: immunochemical localization using light and electron microscopy | journal = The Journal of Histochemistry and Cytochemistry | volume = 39 | issue = 8 | pages = 1001–8 | date = Aug 1991 | pmid = 1856451 | doi = 10.1177/39.8.1856451 | doi-access = free }} |
* {{cite journal | vauthors = Aran JM, Colomer D, Matutes E, Vives-Corrons JL, Franco R | title = Presence of adenosine deaminase on the surface of mononuclear blood cells: immunochemical localization using light and electron microscopy | journal = The Journal of Histochemistry and Cytochemistry | volume = 39 | issue = 8 | pages = 1001–8 | date = Aug 1991 | pmid = 1856451 | doi = 10.1177/39.8.1856451 | doi-access = free }} |
||
* {{cite journal | vauthors = Bielat K, Tritsch GL | title = Ecto-enzyme activity of human erythrocyte adenosine deaminase | journal = Molecular and Cellular Biochemistry | volume = 86 | issue = 2 | pages = 135–42 | date = Apr 1989 | pmid = 2770711 | doi = 10.1007/BF00222613 }} |
* {{cite journal | vauthors = Bielat K, Tritsch GL | title = Ecto-enzyme activity of human erythrocyte adenosine deaminase | journal = Molecular and Cellular Biochemistry | volume = 86 | issue = 2 | pages = 135–42 | date = Apr 1989 | pmid = 2770711 | doi = 10.1007/BF00222613 | s2cid = 20850552 }} |
||
* {{cite journal | vauthors = Hirschhorn R, Tzall S, Ellenbogen A, Orkin SH | title = Identification of a point mutation resulting in a heat-labile adenosine deaminase (ADA) in two unrelated children with partial ADA deficiency | journal = The Journal of Clinical Investigation | volume = 83 | issue = 2 | pages = 497–501 | date = Feb 1989 | pmid = 2783588 | pmc = 303706 | doi = 10.1172/JCI113909 }} |
* {{cite journal | vauthors = Hirschhorn R, Tzall S, Ellenbogen A, Orkin SH | title = Identification of a point mutation resulting in a heat-labile adenosine deaminase (ADA) in two unrelated children with partial ADA deficiency | journal = The Journal of Clinical Investigation | volume = 83 | issue = 2 | pages = 497–501 | date = Feb 1989 | pmid = 2783588 | pmc = 303706 | doi = 10.1172/JCI113909 }} |
||
* {{cite journal | vauthors = Murray JL, Perez-Soler R, Bywaters D, Hersh EM | title = Decreased adenosine deaminase (ADA) and 5'nucleotidase (5NT) activity in peripheral blood T cells in Hodgkin disease | journal = American Journal of Hematology | volume = 21 | issue = 1 | pages = 57–66 | date = Jan 1986 | pmid = 3010705 | doi = 10.1002/ajh.2830210108 }} |
* {{cite journal | vauthors = Murray JL, Perez-Soler R, Bywaters D, Hersh EM | title = Decreased adenosine deaminase (ADA) and 5'nucleotidase (5NT) activity in peripheral blood T cells in Hodgkin disease | journal = American Journal of Hematology | volume = 21 | issue = 1 | pages = 57–66 | date = Jan 1986 | pmid = 3010705 | doi = 10.1002/ajh.2830210108 | s2cid = 25540139 }} |
||
* {{cite journal | vauthors = Wiginton DA, Kaplan DJ, States JC, Akeson AL, Perme CM, Bilyk IJ, Vaughn AJ, Lattier DL, Hutton JJ | title = Complete sequence and structure of the gene for human adenosine deaminase | journal = Biochemistry | volume = 25 | issue = 25 | pages = 8234–44 | date = Dec 1986 | pmid = 3028473 | doi = 10.1021/bi00373a017 }} |
* {{cite journal | vauthors = Wiginton DA, Kaplan DJ, States JC, Akeson AL, Perme CM, Bilyk IJ, Vaughn AJ, Lattier DL, Hutton JJ | title = Complete sequence and structure of the gene for human adenosine deaminase | journal = Biochemistry | volume = 25 | issue = 25 | pages = 8234–44 | date = Dec 1986 | pmid = 3028473 | doi = 10.1021/bi00373a017 }} |
||
* {{cite journal | vauthors = Akeson AL, Wiginton DA, Dusing MR, States JC, Hutton JJ | title = Mutant human adenosine deaminase alleles and their expression by transfection into fibroblasts | journal = The Journal of Biological Chemistry | volume = 263 | issue = 31 | pages = 16291–6 | date = Nov 1988 | pmid = 3182793 | doi = }} |
* {{cite journal | vauthors = Akeson AL, Wiginton DA, Dusing MR, States JC, Hutton JJ | title = Mutant human adenosine deaminase alleles and their expression by transfection into fibroblasts | journal = The Journal of Biological Chemistry | volume = 263 | issue = 31 | pages = 16291–6 | date = Nov 1988 | pmid = 3182793 | doi = }} |
||
* {{cite journal | vauthors = Glader BE, Backer K | title = Elevated red cell adenosine deaminase activity: a marker of disordered erythropoiesis in Diamond-Blackfan anaemia and other haematologic diseases | journal = British Journal of Haematology | volume = 68 | issue = 2 | pages = 165–8 | date = Feb 1988 | pmid = 3348976 | doi = 10.1111/j.1365-2141.1988.tb06184.x }} |
* {{cite journal | vauthors = Glader BE, Backer K | title = Elevated red cell adenosine deaminase activity: a marker of disordered erythropoiesis in Diamond-Blackfan anaemia and other haematologic diseases | journal = British Journal of Haematology | volume = 68 | issue = 2 | pages = 165–8 | date = Feb 1988 | pmid = 3348976 | doi = 10.1111/j.1365-2141.1988.tb06184.x | s2cid = 44789636 }} |
||
* {{cite journal | vauthors = Petersen MB, Tranebjaerg L, Tommerup N, Nygaard P, Edwards H | title = New assignment of the adenosine deaminase gene locus to chromosome 20q13 X 11 by study of a patient with interstitial deletion 20q | journal = Journal of Medical Genetics | volume = 24 | issue = 2 | pages = 93–6 | date = Feb 1987 | pmid = 3560174 | pmc = 1049896 | doi = 10.1136/jmg.24.2.93 }} |
* {{cite journal | vauthors = Petersen MB, Tranebjaerg L, Tommerup N, Nygaard P, Edwards H | title = New assignment of the adenosine deaminase gene locus to chromosome 20q13 X 11 by study of a patient with interstitial deletion 20q | journal = Journal of Medical Genetics | volume = 24 | issue = 2 | pages = 93–6 | date = Feb 1987 | pmid = 3560174 | pmc = 1049896 | doi = 10.1136/jmg.24.2.93 }} |
||
* {{cite journal | vauthors = Orkin SH, Goff SC, Kelley WN, Daddona PE | title = Transient expression of human adenosine deaminase cDNAs: identification of a nonfunctional clone resulting from a single amino acid substitution | journal = Molecular and Cellular Biology | volume = 5 | issue = 4 | pages = 762–7 | date = Apr 1985 | pmid = 3838797 | pmc = 366780 | doi = 10.1128/mcb.5.4.762 }} |
* {{cite journal | vauthors = Orkin SH, Goff SC, Kelley WN, Daddona PE | title = Transient expression of human adenosine deaminase cDNAs: identification of a nonfunctional clone resulting from a single amino acid substitution | journal = Molecular and Cellular Biology | volume = 5 | issue = 4 | pages = 762–7 | date = Apr 1985 | pmid = 3838797 | pmc = 366780 | doi = 10.1128/mcb.5.4.762 }} |
||
Revision as of 01:38, 5 October 2020
| Adenosine/AMP deaminase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
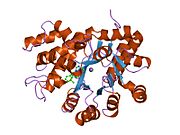
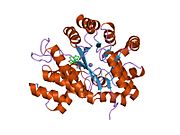
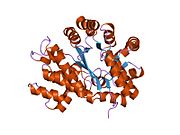
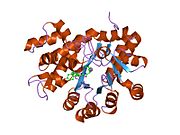
 crystal structure of plasmodium yoelii adenosine deaminase (py02076) | |||||||||
| Identifiers | |||||||||
| Symbol | A_deaminase | ||||||||
| Pfam | PF00962 | ||||||||
| Pfam clan | CL0034 | ||||||||
| InterPro | IPR001365 | ||||||||
| PROSITE | PDOC00419 | ||||||||
| SCOP2 | 1add / SCOPe / SUPFAM | ||||||||
| CDD | cd01320 | ||||||||
| |||||||||
| Adenosine deaminase (editase) domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | A_deamin | ||||||||
| Pfam | PF02137 | ||||||||
| InterPro | IPR002466 | ||||||||
| PROSITE | PDOC00419 | ||||||||
| SCOP2 | 1add / SCOPe / SUPFAM | ||||||||
| |||||||||
| Adenosine/AMP deaminase N-terminal | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | A_deaminase_N | ||||||||
| Pfam | PF08451 | ||||||||
| InterPro | IPR013659 | ||||||||
| |||||||||
Adenosine deaminase (also known as adenosine aminohydrolase, or ADA) is an enzyme (EC 3.5.4.4) involved in purine metabolism. It is needed for the breakdown of adenosine from food and for the turnover of nucleic acids in tissues.
Its primary function in humans is the development and maintenance of the immune system.[5] However, the full physiological role of ADA is not yet completely understood.[6]
Structure
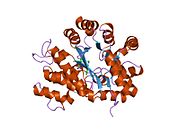
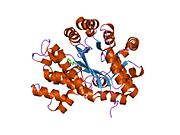
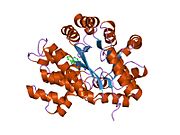
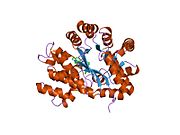
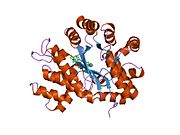
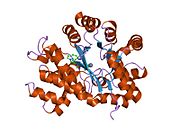
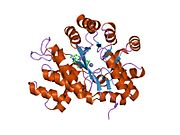
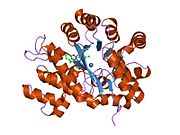
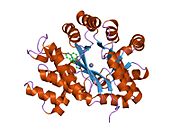
ADA exists in both small form (as a monomer) and large form (as a dimer-complex).[6] In the monomer form, the enzyme is a polypeptide chain,[7] folded into eight strands of parallel α/β barrels, which surround a central deep pocket that is the active site.[5] In addition to the eight central β-barrels and eight peripheral α-helices, ADA also contains five additional helices: residues 19-76 fold into three helices, located between β1 and α1 folds; and two antiparallel carboxy-terminal helices are located across the amino-terminal of the β-barrel.
The ADA active site contains a zinc ion, which is located in the deepest recess of the active site and coordinated by five atoms from His15, His17, His214, Asp295, and the substrate.[5] Zinc is the only cofactor necessary for activity.
The substrate, adenosine, is stabilized and bound to the active site by nine hydrogen bonds.[5] The carboxyl group of Glu217, roughly coplanar with the substrate purine ring, is in position to form a hydrogen bond with N1 of the substrate. The carboxyl group of Asp296, also coplanar with the substrate purine ring, forms hydrogen bond with N7 of the substrate. The NH group of Gly184 is in position to form a hydrogen bond with N3 of the substrate. Asp296 forms bonds both with the Zn2+ ion as well as with 6-OH of the substrate. His238 also hydrogen bonds to substrate 6-OH. The 3'-OH of the substrate ribose forms a hydrogen bond with Asp19, while the 5'-OH forms a hydrogen bond with His17. Two further hydrogen bonds are formed to water molecules, at the opening of the active site, by the 2'-OH and 3'-OH of the substrate.
Due to the recessing of the active site inside the enzyme, the substrate, once bound, is almost completely sequestered from solvent.[5] The surface exposure of the substrate to solvent when bound is 0.5% the surface exposure of the substrate in the free state.
Reactions
ADA irreversibly deaminates adenosine, converting it to the related nucleoside inosine by the substitution of the amino group by a keto group.


Inosine can then be deribosylated (removed from ribose) by another enzyme called purine nucleoside phosphorylase (PNP), converting it to hypoxanthine.
Mechanism of catalysis
The proposed mechanism for ADA-catalyzed deamination is stereospecific addition-elimination via tetrahedral intermediate.[8] By either mechanism, Zn2+ as a strong electrophile activates a water molecule, which is deprotonated by the basic Asp295 to form the attacking hydroxide.[5] His238 orients the water molecule and stabilizes the charge of the attacking hydroxide. Glu217 is protonated to donate a proton to N1 of the substrate.
The reaction is stereospecific due to the location of the zinc, Asp295, and His238 residues, which all face the B-side of the purine ring of the substrate.[5]
Competitive inhibition has been observed for ADA, where the product inosine acts at the competitive inhibitor to enzymatic activity.[9]
Function
ADA is considered one of the key enzymes of purine metabolism.[8] The enzyme has been found in bacteria, plants, invertebrates, vertebrates, and mammals, with high conservation of amino acid sequence.[6] The high degree of amino acid sequence conservation suggests the crucial nature of ADA in the purine salvage pathway.
Primarily, ADA in humans is involved in the development and maintenance of the immune system. However, ADA association has also been observed with epithelial cell differentiation, neurotransmission, and gestation maintenance.[10] It has also been proposed that ADA, in addition to adenosine breakdown, stimulates release of excitatory amino acids and is necessary to the coupling of A1 adenosine receptors and heterotrimeric G proteins.[6] Adenosine deaminase deficiency leads to pulmonary fibrosis,[11] suggesting that chronic exposure to high levels of adenosine can exacerbate inflammation responses rather than suppressing them. It has also been recognized that adenosine deaminase protein and activity is upregulated in mouse hearts that overexpress HIF-1 alpha, which in part explains the attenuated levels of adenosine in HIF-1 alpha expressing hearts during ischemic stress.[12]
Pathology
Some mutations in the gene for adenosine deaminase cause it not to be expressed. The resulting deficiency is one cause of severe combined immunodeficiency (SCID), particularly of autosomal recessive inheritance.[13] Deficient levels of ADA have also been associated with pulmonary inflammation, thymic cell death, and defective T-cell receptor signaling.[14][15]
Conversely, mutations causing this enzyme to be overexpressed are one cause of hemolytic anemia.[16]
There is some evidence that a different allele (ADA2) may lead to autism.[17]
Elevated levels of ADA has also been associated with AIDS.[14][18]
Isoforms
There are 2 isoforms of ADA: ADA1 and ADA2.
- ADA1 is found in most body cells, particularly lymphocytes and macrophages, where it is present not only in the cytosol and nucleus but also as the ecto- form on the cell membrane attached to dipeptidyl peptidase-4 (aka, CD26). ADA1 is involved mostly in intracellular activity, and exists both in small form (monomer) and large form (dimer).[6] The interconversion of small to large forms is regulated by a 'conversion factor' in the lung.[19]
- ADA2 was first identified in human spleen.[20] It was subsequently found in other tissues including the macrophage where it co-exists with ADA1. The two isoforms regulate the ratio of adenosine to deoxyadenosine potentiating the killing of parasites. ADA2 is found predominantly in the human plasma and serum, and exists solely as a homodimer.[21]
Clinical significance
ADA2 is the predominant form present in human blood plasma and is increased in many diseases, particularly those associated with the immune system: for example rheumatoid arthritis, psoriasis, and sarcoidosis. The plasma ADA2 isoform is also increased in most cancers. ADA2 is not ubiquitous but co-exists with ADA1 only in monocytes-macrophages.[citation needed]
Total plasma ADA can be measured using high performance liquid chromatography or enzymatic or colorimetric techniques. Perhaps the simplest system is the measurement of the ammonia released from adenosine when broken down to inosine. After incubation of plasma with a buffered solution of adenosine the ammonia is reacted with a Berthelot reagent to form a blue colour which is proportionate to the amount of enzyme activity. To measure ADA2, erythro-9-(2-hydroxy-3-nonyl) adenine (EHNA) is added prior to incubation so as to inhibit the enzymatic activity of ADA1.[20] It is the absence of ADA1 that causes SCID.
ADA can also be used in the workup of lymphocytic pleural effusions or peritoneal ascites, in that such specimens with low ADA levels essentially excludes tuberculosis from consideration.[22]
Tuberculosis pleural effusions can now be diagnosed accurately by increased levels of pleural fluid adenosine deaminase, above 40 U per liter.[23]
Cladribine and Pentostatin are anti-neoplastic agents used in the treatment of hairy cell leukemia; their mechanism of action is inhibition of adenosine deaminase.
See also
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000196839 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000017697 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b c d e f g Wilson DK, Rudolph FB, Quiocho FA (May 1991). "Atomic structure of adenosine deaminase complexed with a transition-state analog: understanding catalysis and immunodeficiency mutations". Science. 252 (5010): 1278–1284. doi:10.1126/science.1925539. PMID 1925539.
- ^ a b c d e Cristalli G, Costanzi S, Lambertucci C, Lupidi G, Vittori S, Volpini R, Camaioni E (Mar 2001). "Adenosine deaminase: functional implications and different classes of inhibitors". Medicinal Research Reviews. 21 (2): 105–128. doi:10.1002/1098-1128(200103)21:2<105::AID-MED1002>3.0.CO;2-U. PMID 11223861.
- ^ Daddona PE, Kelley WN (Jan 1977). "Human adenosine deaminase. Purification and subunit structure". The Journal of Biological Chemistry. 252 (1): 110–115. PMID 13062.
- ^ a b Losey HC, Ruthenburg AJ, Verdine GL (Jan 2006). "Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA". Nature Structural & Molecular Biology. 13 (2): 153–159. doi:10.1038/nsmb1047. PMID 16415880. S2CID 34848284.
- ^ Saboury AA, Divsalar A, Jafari GA, Moosavi-Movahedi AA, Housaindokht MR, Hakimelahi GH (May 2002). "A product inhibition study on adenosine deaminase by spectroscopy and calorimetry". Journal of Biochemistry and Molecular Biology. 35 (3): 302–305. doi:10.5483/BMBRep.2002.35.3.302. PMID 12297022.
- ^ Moriwaki Y, Yamamoto T, Higashino K (Oct 1999). "Enzymes involved in purine metabolism--a review of histochemical localization and functional implications". Histology and Histopathology. 14 (4): 1321–1340. PMID 10506947.
- ^ Blackburn MR (2003). "Too much of a good thing: adenosine overload in adenosine-deaminase-deficient mice". Trends in Pharmacological Sciences. 24 (2): 66–70. doi:10.1016/S0165-6147(02)00045-7. PMID 12559769.
- ^ Wu J, Bond C, Chen P, Chen M, Li Y, Shohet RV, Wright G (2015). "HIF-1α in the heart: remodeling nucleotide metabolism". Journal of Molecular and Cellular Cardiology. 82: 194–200. doi:10.1016/j.yjmcc.2015.01.014. PMC 4405794. PMID 25681585.
- ^ Sanchez JJ, Monaghan G, Børsting C, Norbury G, Morling N, Gaspar HB (May 2007). "Carrier frequency of a nonsense mutation in the adenosine deaminase (ADA) gene implies a high incidence of ADA-deficient severe combined immunodeficiency (SCID) in Somalia and a single, common haplotype indicates common ancestry". Annals of Human Genetics. 71 (Pt 3): 336–47. doi:10.1111/j.1469-1809.2006.00338.x. PMID 17181544. S2CID 34850391.
- ^ a b Blackburn MR, Kellems RE (2005). "Adenosine deaminase deficiency: metabolic basis of immune deficiency and pulmonary inflammation". Advances in Immunology. 86: 1–41. doi:10.1016/S0065-2776(04)86001-2. ISBN 9780120044863. PMID 15705418.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Apasov SG, Blackburn MR, Kellems RE, Smith PT, Sitkovsky MV (Jul 2001). "Adenosine deaminase deficiency increases thymic apoptosis and causes defective T cell receptor signaling". The Journal of Clinical Investigation. 108 (1): 131–141. doi:10.1172/JCI10360. PMC 209335. PMID 11435465.
- ^ Chottiner EG, Cloft HJ, Tartaglia AP, Mitchell BS (Mar 1987). "Elevated adenosine deaminase activity and hereditary hemolytic anemia. Evidence for abnormal translational control of protein synthesis". The Journal of Clinical Investigation. 79 (3): 1001–5. doi:10.1172/JCI112866. PMC 424261. PMID 3029177.
- ^ Persico AM, Militerni R, Bravaccio C, Schneider C, Melmed R, Trillo S, Montecchi F, Palermo MT, Pascucci T, Puglisi-Allegra S, Reichelt KL, Conciatori M, Baldi A, Keller F (Dec 2000). "Adenosine deaminase alleles and autistic disorder: case-control and family-based association studies". American Journal of Medical Genetics. 96 (6): 784–90. doi:10.1002/1096-8628(20001204)96:6<784::AID-AJMG18>3.0.CO;2-7. PMID 11121182.
- ^ Cowan MJ, Brady RO, Widder KJ (Feb 1986). "Elevated erythrocyte adenosine deaminase activity in patients with acquired immunodeficiency syndrome". Proceedings of the National Academy of Sciences of the United States of America. 83 (4): 1089–1091. doi:10.1073/pnas.83.4.1089. PMC 323016. PMID 3006027.
- ^ Schrader WP, Stacy AR (Sep 1977). "Purification and subunit structure of adenosine deaminase from human kidney". The Journal of Biological Chemistry. 252 (18): 6409–6415. PMID 893413.
- ^ a b Schrader WP, Pollara B, Meuwissen HJ (Jan 1978). "Characterization of the residual adenosine deaminating activity in the spleen of a patient with combined immunodeficiency disease and adenosine deaminase deficiency". Proceedings of the National Academy of Sciences of the United States of America. 75 (1): 446–50. doi:10.1073/pnas.75.1.446. PMC 411266. PMID 24216.
- ^ Zavialov AV, Engström A (Oct 2005). "Human ADA2 belongs to a new family of growth factors with adenosine deaminase activity". The Biochemical Journal. 391 (Pt 1): 51–57. doi:10.1042/BJ20050683. PMC 1237138. PMID 15926889.
- ^ Jiménez Castro D, Díaz Nuevo G, Pérez-Rodríguez E, Light RW (2003). "Diagnostic value of adenosine deaminase in nontuberculous lymphocytic pleural effusions" (PDF). Eur. Respir. J. 21 (2): 220–4. doi:10.1183/09031936.03.00051603. PMID 12608433. S2CID 10703687.
- ^ Brunicardi, F.; Andersen, Dana; Billiar, Timothy; Dunn, David; Hunter, John; Pollock, Raphael E. (2005). "Chapter 18, question 16". Schwartz's principles of surgery (8th ed.). New York: McGraw-Hill Professional. ISBN 978-0071410908.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help)
Further reading
- da Cunha JG (1992). "[Adenosine deaminase. A pluridisciplinary enzyme]". Acta Médica Portuguesa. 4 (6): 315–23. PMID 1807098.
- Franco R, Casadó V, Ciruela F, Saura C, Mallol J, Canela EI, Lluis C (Jul 1997). "Cell surface adenosine deaminase: much more than an ectoenzyme". Progress in Neurobiology. 52 (4): 283–94. doi:10.1016/S0301-0082(97)00013-0. PMID 9247966. S2CID 40318396.
- Valenzuela A, Blanco J, Callebaut C, Jacotot E, Lluis C, Hovanessian AG, Franco R (1997). "HIV-1 envelope gp120 and viral particles block adenosine deaminase binding to human CD26". Advances in Experimental Medicine and Biology. 421: 185–92. doi:10.1007/978-1-4757-9613-1_24. ISBN 978-1-4757-9615-5. PMID 9330696.
- Moriwaki Y, Yamamoto T, Higashino K (Oct 1999). "Enzymes involved in purine metabolism--a review of histochemical localization and functional implications". Histology and Histopathology. 14 (4): 1321–40. PMID 10506947.
- Hirschhorn R (1993). "Identification of two new missense mutations (R156C and S291L) in two ADA- SCID patients unusual for response to therapy with partial exchange transfusions". Human Mutation. 1 (2): 166–8. doi:10.1002/humu.1380010214. PMID 1284479. S2CID 44617309.
- Berkvens TM, van Ormondt H, Gerritsen EJ, Khan PM, van der Eb AJ (Aug 1990). "Identical 3250-bp deletion between two AluI repeats in the ADA genes of unrelated ADA-SCID patients". Genomics. 7 (4): 486–90. doi:10.1016/0888-7543(90)90190-6. PMID 1696926.
- Aran JM, Colomer D, Matutes E, Vives-Corrons JL, Franco R (Aug 1991). "Presence of adenosine deaminase on the surface of mononuclear blood cells: immunochemical localization using light and electron microscopy". The Journal of Histochemistry and Cytochemistry. 39 (8): 1001–8. doi:10.1177/39.8.1856451. PMID 1856451.
- Bielat K, Tritsch GL (Apr 1989). "Ecto-enzyme activity of human erythrocyte adenosine deaminase". Molecular and Cellular Biochemistry. 86 (2): 135–42. doi:10.1007/BF00222613. PMID 2770711. S2CID 20850552.
- Hirschhorn R, Tzall S, Ellenbogen A, Orkin SH (Feb 1989). "Identification of a point mutation resulting in a heat-labile adenosine deaminase (ADA) in two unrelated children with partial ADA deficiency". The Journal of Clinical Investigation. 83 (2): 497–501. doi:10.1172/JCI113909. PMC 303706. PMID 2783588.
- Murray JL, Perez-Soler R, Bywaters D, Hersh EM (Jan 1986). "Decreased adenosine deaminase (ADA) and 5'nucleotidase (5NT) activity in peripheral blood T cells in Hodgkin disease". American Journal of Hematology. 21 (1): 57–66. doi:10.1002/ajh.2830210108. PMID 3010705. S2CID 25540139.
- Wiginton DA, Kaplan DJ, States JC, Akeson AL, Perme CM, Bilyk IJ, Vaughn AJ, Lattier DL, Hutton JJ (Dec 1986). "Complete sequence and structure of the gene for human adenosine deaminase". Biochemistry. 25 (25): 8234–44. doi:10.1021/bi00373a017. PMID 3028473.
- Akeson AL, Wiginton DA, Dusing MR, States JC, Hutton JJ (Nov 1988). "Mutant human adenosine deaminase alleles and their expression by transfection into fibroblasts". The Journal of Biological Chemistry. 263 (31): 16291–6. PMID 3182793.
- Glader BE, Backer K (Feb 1988). "Elevated red cell adenosine deaminase activity: a marker of disordered erythropoiesis in Diamond-Blackfan anaemia and other haematologic diseases". British Journal of Haematology. 68 (2): 165–8. doi:10.1111/j.1365-2141.1988.tb06184.x. PMID 3348976. S2CID 44789636.
- Petersen MB, Tranebjaerg L, Tommerup N, Nygaard P, Edwards H (Feb 1987). "New assignment of the adenosine deaminase gene locus to chromosome 20q13 X 11 by study of a patient with interstitial deletion 20q". Journal of Medical Genetics. 24 (2): 93–6. doi:10.1136/jmg.24.2.93. PMC 1049896. PMID 3560174.
- Orkin SH, Goff SC, Kelley WN, Daddona PE (Apr 1985). "Transient expression of human adenosine deaminase cDNAs: identification of a nonfunctional clone resulting from a single amino acid substitution". Molecular and Cellular Biology. 5 (4): 762–7. doi:10.1128/mcb.5.4.762. PMC 366780. PMID 3838797.
- Valerio D, Duyvesteyn MG, Dekker BM, Weeda G, Berkvens TM, van der Voorn L, van Ormondt H, van der Eb AJ (Feb 1985). "Adenosine deaminase: characterization and expression of a gene with a remarkable promoter". The EMBO Journal. 4 (2): 437–43. doi:10.1002/j.1460-2075.1985.tb03648.x. PMC 554205. PMID 3839456.
- Bonthron DT, Markham AF, Ginsburg D, Orkin SH (Aug 1985). "Identification of a point mutation in the adenosine deaminase gene responsible for immunodeficiency". The Journal of Clinical Investigation. 76 (2): 894–7. doi:10.1172/JCI112050. PMC 423929. PMID 3839802.
- Daddona PE, Shewach DS, Kelley WN, Argos P, Markham AF, Orkin SH (Oct 1984). "Human adenosine deaminase. cDNA and complete primary amino acid sequence". The Journal of Biological Chemistry. 259 (19): 12101–6. PMID 6090454.
- Valerio D, Duyvesteyn MG, Meera Khan P, Geurts van Kessel A, de Waard A, van der Eb AJ (Nov 1983). "Isolation of cDNA clones for human adenosine deaminase". Gene. 25 (2–3): 231–40. doi:10.1016/0378-1119(83)90227-5. PMID 6198240.
External links
- ADA human gene location in the UCSC Genome Browser.
- ADA human gene details in the UCSC Genome Browser.
- PDBe-KB provides an overview of all the structure information available in the PDB for Human Adenosine deaminase
- PDBe-KB provides an overview of all the structure information available in the PDB for Mouse Adenosine deaminase