Talk:Variants of SARS-CoV-2
| This is the talk page for discussing improvements to the Variants of SARS-CoV-2 article. This is not a forum for general discussion of the article's subject. |
Article policies
|
| Find medical sources: Source guidelines · PubMed · Cochrane · DOAJ · Gale · OpenMD · ScienceDirect · Springer · Trip · Wiley · TWL |
| Archives: 1Auto-archiving period: 12 months |
| The contentious topics procedure applies to this page. This page is related to COVID-19, broadly construed, which has been designated as a contentious topic. Editors who repeatedly or seriously fail to adhere to the purpose of Wikipedia, any expected standards of behaviour, or any normal editorial process may be blocked or restricted by an administrator. Editors are advised to familiarise themselves with the contentious topics procedures before editing this page. |
WikiProject COVID-19 consensus WikiProject COVID-19 aims to add to and build consensus for pages relating to COVID-19. They have so far discussed items listed below. Please discuss proposed improvements to them at the project talk page.
To ensure you are viewing the current list, you may wish to . |
| This article has not yet been rated on Wikipedia's content assessment scale. It is of interest to multiple WikiProjects. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Please add the quality rating to the {{WikiProject banner shell}} template instead of this project banner. See WP:PIQA for details.
Please add the quality rating to the {{WikiProject banner shell}} template instead of this project banner. See WP:PIQA for details.
Please add the quality rating to the {{WikiProject banner shell}} template instead of this project banner. See WP:PIQA for details.
Please add the quality rating to the {{WikiProject banner shell}} template instead of this project banner. See WP:PIQA for details.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 | Ideal sources for Wikipedia's health content are defined in the guideline Wikipedia:Identifying reliable sources (medicine) and are typically review articles. Here are links to possibly useful sources of information about Variants of SARS-CoV-2.
|
| Text and/or other creative content from this version of Severe acute respiratory syndrome coronavirus 2#Strains and variants was copied or moved into Severe acute respiratory syndrome coronavirus 2 variants with this edit. The former page's history now serves to provide attribution for that content in the latter page, and it must not be deleted as long as the latter page exists. |
| Text and/or other creative content from this version of 501.V2 Variant was copied or moved into Severe acute respiratory syndrome coronavirus 2 variants with this edit. The former page's history now serves to provide attribution for that content in the latter page, and it must not be deleted as long as the latter page exists. |
| Text and/or other creative content from this version of Cluster 5 was copied or moved into Severe acute respiratory syndrome coronavirus 2 variants with this edit. The former page's history now serves to provide attribution for that content in the latter page, and it must not be deleted as long as the latter page exists. |
| Text and/or other creative content from this version of VOC-202012/01 was copied or moved into Severe acute respiratory syndrome coronavirus 2 variants with this edit. The former page's history now serves to provide attribution for that content in the latter page, and it must not be deleted as long as the latter page exists. |
| This is the talk page for discussing improvements to the Variants of SARS-CoV-2 article. This is not a forum for general discussion of the article's subject. |
Article policies
|
| Find medical sources: Source guidelines · PubMed · Cochrane · DOAJ · Gale · OpenMD · ScienceDirect · Springer · Trip · Wiley · TWL |
| Archives: 1Auto-archiving period: 12 months |
New variant P.1
There's a new variant P.1 (descendent of B.1.1.28) that's getting some attention and may reach the level of notability to deserve a section in this article.
- @billhanage (Jan 12, 2021). "There is a new 'variant' clearly identified today, P.1. And it is worth saying a little about what we have learned about it, and from the variants we have identified so far https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 1/quite_a_few" (Tweet) – via Twitter.
citing
- Nuno R. Faria (Jan 12, 2021). "Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings". Virological.
(It may be what is referred to above as Talk:Variants of SARS-CoV-2/Archive 1#Japan/Brazil variant, but maybe not?) jhawkinson (talk) 14:04, 13 January 2021 (UTC)
- It does definitely seem of interest considering the RBD mutations, but I don't think there has has been enough reported of it to introduce it yet. I suspect there will be very soon, though. Regarding your last sentence, they're the same. https://cov-lineages.org/lineages/lineage_B.1.1.28.html says:
[P.1 is the alias] of B.1.1.28.1, Brazilian lineage with a number of spike mutaions with likely functional significance E484K, K417T, and N501Y. Described in https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586
. —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 15:18, 13 January 2021 (UTC)- That reminds me that perhaps https://cov-lineages.org should be an EL on this page. jhawkinson (talk) 15:36, 13 January 2021 (UTC)
- I suspect there is some confusion going on both on this page and on Lineage B.1.1.248. Specifically, it appears there are 2 widely discussed variants thought to be originated in Brazil. One is referred to as P.1, B.1.1.28.1, and 20J/501Y.V3. It has both N501Y and E484K mutations. Another is referred to as P.2, B.1.1.248, and 20B/S.484K. According to "COG-UK report on SARS-CoV-2 Spike mutations of interest in the UK 15th January 2021" (PDF). P.2 has E484K but not N501Y. Both this article and Lineage B.1.1.248 seem to combine information about P.1 and P.2 assuming it is the same lineage. However, "National Institute of Infectious Diseases report "New Variant Strainof SARS-CoV-2Identified in Travelers from Brazil"" (PDF). seems to use the name B.1.1.248 for P.1 instead of P.2 adding to the confusion. I'm trying to verify the above claims to resolve the confusion. Vikasatkin (talk) 18:13, 16 January 2021 (UTC)
- @Vikasatkin: I regret that, as regards 'Variants_of_SARS-CoV-2 / $ B.1.1.248', lack of clarity is largely my fault. To be explicit, the first sentence of the second para of Lineage B.1.1.248 could perhaps usefully have read
- "A preprint of a paper by Carolina M Voloch et al. identified a novel lineage of SARS-CoV-2 in circulation in Brazil, 'B.1.1.248', which originated from B.1.1.28." -and similarly, later in that para, 'novel variant lineage' could have '(B.1.1.248)' added to remove all possibility of doubt. I must re-read the source for further distinctions...
- Use of P1 and P2 is perhaps best kept back for more detail in the main article. Yadsalohcin (talk) 22:20, 16 January 2021 (UTC)
'A701B'(?)
Little seems to be forthcoming re mutation 'A701B'(sic) beyond the two existing refs from The Straits Times, 23 December 2020 and GMA News, 27 December 2020. However, I note that there is a more technical source (mirrored on the FB page for Noor Hisham Abdullah, who is quoted as the source for the Straits Times article) which refers to 'A701V' in very similar terms.
- On 25 December 2020, the organisation 'Kementerian Kesihatan Malaysia/ covid-19 Malaysia' wrote: "A701V mutation; ... was first found among 22 SARS-CoV-2 sequences from the Benteng LD clusters and subsequently passed on to the vast majority of third wave clusters in Peninsular Malaysia and Sabah."[1]
cf.
- The Straits Times (currently ref 46) wrote: " Malaysia's Health Ministry said on Wednesday (Dec 23) it has identified a new Covid-19 strain in the country from samples taken in Sabah. The ministry's director-general, Tan Sri Dr Noor Hisham Abdullah, said it is still unknown whether the strain - dubbed the "A701B" mutation - is more infectious than usual. "It is similar to a strain found in South Africa, Australia and the Netherlands," he said at his daily news conference."[2]
- GMA News (currently ref 47) wrote: "reports that a new coronavirus disease 2019 (COVID-19) variant has been detected in neighboring Sabah, Malaysia... Malaysia’s Health Ministry has dubbed the new variant as the "A701B" mutation"[3]
It seems to me that we have the same thing under discussion, but with its name potentially misreported (possibly via a mis-hearing?)
I am therefore minded to change the existing section "A701B" to "A701V" retaining A701B in the text but appending '(sic)', and add the following:
- On 25 December 2020, the organisation 'Kementerian Kesihatan Malaysia/ covid-19 Malaysia' described a mutation A701V as circulating and present in 85% of cases (D614G was present in 100% of cases) in Malaysia.[1]
- This (A701V) mutation has the amino acid Alanine substituted by Valine at codon position 701 in the spike protein. The source states that globally, South Africa, Australia, Netherlands and England also reported A701V at about the same time as Malaysia.[1] In GISAID, the prevalence of this mutation is found to be about 0.18%. of cases.[1]
Yadsalohcin (talk) 19:45, 15 January 2021 (UTC)
- I've removed this mutation from the summary of variants until further information can be found. No information about a specific variant has been found and there are conflicting reports about the mutation itself. Side note: For those that are unaware, A701B indicated an alanine (GCN) to aspartic acid (GAY) or asparagine (AAY) mutation, whereas, as you have written, A701V indicates alanine (GCN) to valine (GUN). ArcMachaon (talk) 02:40, 16 January 2021 (UTC)
- And I've commented the A701V. I think we should wait till this gets clarified by an RS. —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 03:41, 16 January 2021 (UTC)
References
- ^ a b c d The current situation and Information on the Spike protein mutation of Covid-19 in Malaysia 25 December 2020 Kementerian Kesihatan Malaysia/ covid-19 Malaysia/ covid-19.moh.gov.my accessed 15 January 2021
- ^ "Malaysia identifies new Covid-19 strain, similar to one found in 3 other countries". The Straits Times. 23 December 2020. Retrieved 10 January 2021.
- ^ "Duterte says Sulu seeking help after new COVID-19 variant detected in nearby Sabah, Malaysia". GMA News. 27 December 2020. Retrieved 10 January 2021.
COH.20G/501Y
Researchers Discover New Variant of COVID-19 Virus in Columbus, Ohio I don't know enough to be able to tell if this is already generally covered in the article, specifically regarding the "20G" ID. Mapsax (talk) 00:23, 16 January 2021 (UTC)
- Hi Mapsax, thanks for this, for the moment the 'obvious place' seems to me to be a further addition to the sub section Notable mutations/ N501Y... only time will tell what we should eventually do with this information. Yadsalohcin (talk) 01:07, 16 January 2021 (UTC)
Other variants in the U.S. and Canada
At the risk of Ohio becoming WP:UNDUE, I wanted to mention that I've heard of other variants in the mainstream media throughout North America – as I type this, they've been reported in New York, Louisiana, Texas, and Ontario – but I don't know for sure if those are existing variants (I do know that reports call the variant in California the "UK variant") and wouldn't know which sources would be accepted as RS in this context. Mapsax (talk) 23:39, 16 January 2021 (UTC)
Roman numerals in headings
Roman numerals, or any other number should not be used in the headings for several reasons:
- The headings are already numbered in the contents box and do not need to be renumbered
- The numbering system may be mistaken for something relevant, such as an official designation or numbering system
- It is unwieldy and not particularly aesthetic
- It goes against WP:HEADING, which states headings should not be numbered etc.
Therefore, I have removed the numbering from the article. ArcMachaon (talk) 02:45, 16 January 2021 (UTC)
- thanks for post--Ozzie10aaaa (talk) 20:11, 15 February 2021 (UTC)
Summary table
Currently, without the four pipes seperating "Date" and "Transmissibility", I am seeing the summary table like this: https://imgur.com/a/ddI6Ot6 (notice position of "Transmissibility", "Virulence"; and "Antigenicity"), both on my desktop and laptop. I realise some people might be seeing it correctly (I also see it correctly in the mobile version). Anyone know how to fix this? —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 23:38, 16 January 2021 (UTC)
- thank you--Ozzie10aaaa (talk) 20:11, 15 February 2021 (UTC)
Nonspecific incoming redirects
There are a bunch of non-specific incoming redirects. They need to be handled with hatnotes.
{{redirect|Coronavirus variant|variations of coronavirus|Coronavirus}}{{redirect|New coronavirus variant|recent variations of coronavirus|Novel coronavirus}}{{redirect|New Covid|COVID nomenclature|wikt:COVID{{!}}Wiktionary]]|recently identified coronavirus diseases|Coronavirus disease}}
Clearly, this article doses not cover all coronaviruses, so it should carry hatnotes to handle these incredibly generic terms.
-- 70.31.205.108 (talk) 14:43, 18 January 2021 (UTC)
WIV04/2019
Shouldn't the "original" strain(s) also have a section? (the two major variants identified in Wuhan in January) -- 70.31.205.108 (talk) 14:47, 18 January 2021 (UTC)
- The original strain is covered at Severe acute respiratory syndrome coronavirus 2. —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 14:58, 19 January 2021 (UTC)
- This article is "variants of", so it would seem logical to have a section for that (the original variants), with a short summary, and a redirect to the base article -- 70.31.205.108 (talk) 08:04, 21 January 2021 (UTC)
- A sequence zero/reference sequence cannot be a variant as it is not different from the reference sequence, as per PMC 3535543:
Van Regenmortel defined a virus variant as an isolate or a set of isolates whose genomic (consensus) sequence(s) differ(s) from that of a reference virus.
—Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 08:11, 21 January 2021 (UTC)- The reference sequence is itself a variant in the spectrum of all variations. Though not such within the field's scientific literature, it is such in as a logical consequence, since the original virus is never the reference sequence, as the sequence that is referenced is inevitably a descendant of the original, sometimes a much later one. -- 70.31.205.108 (talk) 03:50, 25 January 2021 (UTC)
- A sequence zero/reference sequence cannot be a variant as it is not different from the reference sequence, as per PMC 3535543:
- This article is "variants of", so it would seem logical to have a section for that (the original variants), with a short summary, and a redirect to the base article -- 70.31.205.108 (talk) 08:04, 21 January 2021 (UTC)
"COVID-19 variant"
Various incoming redirects like COVID-19 variant point here, shouldn't they point to coronavirus disease 2019? The various different presentations of the disease are covered in the main disease article.
If they remain pointed here, there needs to be a hatnote for it, like:
{{redirect|COVID-19 variant|variations in the course of COVID-19 disease progression|Coronavirus disease 2019}}
-- 70.31.205.108 (talk) 14:53, 18 January 2021 (UTC)
- You can do that if you want to, but "COVID-19 variant" should redirect to this article: even though technically incorrect, people who don't know much about virology might type in "COVID-19 variant", expecting something like a list of variants of SARS-CoV-2. —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 14:58, 19 January 2021 (UTC)
L452R in California
I have been seeing multiple articles about L452R variant in Santa Clara County, CA. News article 1, News article 2,County press release. Should this get a section on this page? --Gimelthedog (talk) 21:30, 18 January 2021 (UTC)
- I say we wait a bit. There seems to quite a bit on L452R (it's mentioned in Cell as a mutation that is
markedly resistant to some mAbs
), [1] but it's only one mutation and has been around since at least May of last year[2]—maybe even February (per "Methods" on p.1). Furthermore, so far all reports have been on transmission with L452R in California; I personally think we should wait for it spread to other countries before writing about it because of the risk of becoming WP:UNDUE. —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 14:58, 19 January 2021 (UTC)
- Also referred to in the New York Times as CAL.20C[3]; there's little reason to wait before adding a variant, people will come here to look for it — it's definitely a variant and that should be enough for notability. The level of information and its import will of course change over time, and we'll change the article appropriately. jhawkinson (talk) 23:18, 19 January 2021 (UTC)
- Sorry I didn't check the talk page before adding the CAL.20C variant section... I tend to agree with jhawkinson in that there's no real reason to wait before adding this variant to the page. I am a little annoyed with the Cedars-Sinai team however... In their press release, they claimed that there is a preprint on MedRxiv.org [4] but I can't seem to find any 2021 submissions by any of the coauthors listed in the press release. Mar2194 (talk) 00:40, 20 January 2021 (UTC)
- The claim in the press release is that the preprint was "submitted" to medRxiv. You might need to wait a bit before it gets approved by medRxiv and made public. Could be longer if medRxiv rejects the first submission. In any case, I don't think Wikipedia allows medRxiv references. Also, the URL for the press release has gone bad, but I found it cached on Google. Jaredroach (talk) 03:56, 25 February 2021 (UTC)
- Sorry I didn't check the talk page before adding the CAL.20C variant section... I tend to agree with jhawkinson in that there's no real reason to wait before adding this variant to the page. I am a little annoyed with the Cedars-Sinai team however... In their press release, they claimed that there is a preprint on MedRxiv.org [4] but I can't seem to find any 2021 submissions by any of the coauthors listed in the press release. Mar2194 (talk) 00:40, 20 January 2021 (UTC)
References
- ^ https://www.cell.com/cell/pdf/S0092-8674%2820%2930877-1.pdf, p.1
- ^ https://www.who.int/bulletin/volumes/98/7/20-253591/en/, figure 3. It's hard to spot, but you can go to doi:10.2471/BLT.20.253591 and use CTRL + F to find it along the second row of figure 3 on p. 6.
- ^ Carl Zimmer (January 19, 2021). "New California Variant May Be Driving Virus Surge There, Study Suggests". Event occurs at 5:24 p.m. ET.
Researchers found that the variant originated in California and showed up in more than half of samples tested last week by researchers in Los Angeles.
- ^ https://www.cedars-sinai.org/newsroom/local-covid-19-strain-found-in-over-one-third-of-los-angeles-patients/
Nomenclature table
The nomenclature table is a bit of a mess. That's not really our fault: the nomenclature in general is "a bloody mess at the moment", as one scientist put it.[1] To avoid further confusion—and because we've already nosedived into WP:OR territory—I suggest deleting the table for now until a secondary source compiles the nomenclatures, like Alm et al. did (although that table, of course, is outdated). —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 14:01, 20 January 2021 (UTC)
- Hello there, B-i-C, yes, the nomenclature table is undoubtedly a bit of a mess, for the reason stated, and because virusses in their evolutionary development don't necessarily choose to accommodate themselves to the layout on a table. However, with one exception (see below) it verifiably follows the reliable sources available to us at the moment and I would contend is a potentially helpful summary of variant names (and with 3 different schemes available there's plenty of room for getting lost).
- As a demonstration of its worthiness, it for example draws attention to the fact that I had over eagerly added CAL.20C to it when in fact we have so far only two news media sources for CAL.20C on this page, which (I think, on closer inspection) do not define its position within any of the naming schemes- its name in this sense would lead one to assume (sorry!) that it belonged on the 20C line, whereas the 'Nextstrain-amendation' source mentions 20G: (derived from 20C... 'concentrated in the United States') which might suggest that it should be with 20G, so, time to cancel said speculation and remove CAL.20C from the table. The table is then all fully supported once more by the refs. It will be interesting to see what more we learn about CAL.20C in the coming weeks. Yadsalohcin (talk) 15:19, 20 January 2021 (UTC)
- Thank you Yadsalohcin! I was going to add it to the table as well but since the Cedars-Sinai preprint is mythological at this point and there are no other references in the literature, I held off. "Interesting" is one way to put it... Mar2194 (talk) 16:31, 20 January 2021 (UTC)
Linkouts to resources
Human genes and protein pages link to various interactive viewers where one can see data about the gene graphically, on the chromosome, 3D structures, protein data etc. For Covid variants there is currently no linkout on this page. I work for one of these websites (UCSC Genome Browser) and I'm wondering if I should start linking to the info graphics that we generate, with mutation frequency, where they are in the SARS-CoV-2 genome, what the protein models on this mutation are, etc. One place could be the nomenclature table. There are a few other resources this page could link to, Nextstrain for example, they may have a URL scheme where the viewer zooms directly onto a mutation and one can get the map of where it currently is found. Maximilianh (talk) 09:35, 21 January 2021 (UTC)
- Hi Maximilianh, going beyond this basic table which cross-references nomenclatures, there are several obvious possible extras- in particular:
- date and location first detected,
- date of sample set (as there are often delays testing and reporting) and region of 'current' greatest prevalence
* principal mutations (but this is now taken care-of in the table in the summary section)- -Further to these, we have a number of refs and a footnote link from the table, so I don't see why we shouldn't also include refs linking to various external viewers and map pages... seems a potentially valuable hub for information- I'd love to see them here. Yadsalohcin (talk) 00:55, 22 January 2021 (UTC)
Fresh preprint about South African variant and antibody escape
Do this worth mentioning in article? --Unxed (talk) 18:52, 21 January 2021 (UTC)
- Not sure. It's a preprint, so we might wanna wait till peer-reviewed version is available. —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 20:03, 21 January 2021 (UTC)
- We mention a couple of Preprints already; I would have thought that provided its 'preprint nature' is clearly stated, mention of the likely upshot of this study would seem in order (tho' maybe in the 501.V2 variant main article first?) Yadsalohcin (talk) 01:00, 22 January 2021 (UTC)
- Added link to 501.V2 talks page, so it could be discussed there also --Unxed (talk) 14:18, 22 January 2021 (UTC)
- We mention a couple of Preprints already; I would have thought that provided its 'preprint nature' is clearly stated, mention of the likely upshot of this study would seem in order (tho' maybe in the 501.V2 variant main article first?) Yadsalohcin (talk) 01:00, 22 January 2021 (UTC)
Article's content does not meet its title
The title of the article is "Variants of SARS-CoV-2". Yet the avowed content is "[those] that are or have been believed to be of particular importance". This article needs an expert to explain the criteria for importance and why unimportant variants can be omitted. It would be useful for that expert to explain whether the rate of all new variants is consistent with that of other viruses or whether the preventive measures we have adopted are part of an arms race with the virus. Robert P. O'Shea (talk) 05:48, 23 January 2021 (UTC)
- I do not entirely disagree, but it's only to be expected that an article about "Variants of X" talks about variants of X in general, and then discusses notable variants of X further. Establishing strict criteria for notability, other than wikipedia's general and rather fluffy criteria, is probably not possible at this point. I think the lead is sufficiently clear about this, saying "some are or have been believed to be of particular importance".--Nø (talk) 12:15, 23 January 2021 (UTC)
- From https://www.who.int/bulletin/volumes/98/7/20-253591/en/ (used in the article): "We detected in total 65776 variants with 5775 distinct variants". Considering these data are from at least May 2020, maybe even February, and that they obviously only looked at a limited number of genomes, it is only reasonable to suggest that there are millions of variants. We can't write about them all. We write about variants that have been reported widely in media (whether that be popular or specialised) and are determined to be of importance in one way or another by these sources. —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 18:06, 27 January 2021 (UTC)
E484K
Is this RS? https://www.biorxiv.org/content/10.1101/2020.12.31.425021v1.article-info Charles Juvon (talk) 21:32, 24 January 2021 (UTC)
- Also, see: https://science.sciencemag.org/content/371/6527/329 Charles Juvon (talk) 22:20, 24 January 2021 (UTC)
Table Colors
Could we swap red and green background color in the table for a more intuitive representation like "red=danger, green=irrelevant" ? --jOERG 17:20, 28 January 2021 (UTC) — Preceding unsigned comment added by Joerg rw (talk • contribs) done --jOERG 18:48, 28 January 2021 (UTC) — Preceding unsigned comment added by Joerg rw (talk • contribs)
Garmisch Variant turns out to be B.1.1.134, not novel
Turns out the unusual COVID variant that was detected in Garmisch-Patenkirchen are not novel, but belong to the sporadically, previously seen B.1.1.134 lineage. Unfortunately, I only have this German source on hand: https://www.br.de/nachrichten/bayern/corona-variante-aus-garmisch-partenkirchen-ist-entschluesselt,SMjr7L1 134.3.255.250 (talk) 13:46, 31 January 2021 (UTC)
- thank you for post--Ozzie10aaaa (talk) 13:25, 22 February 2021 (UTC)
Remove the ticks and crosses
Please remove the {{ya}} and {{na}} from the summary table. A tick usually means yes, and the x cross usually means no, and it took me some time before realising that this is not what is meant in the article. For example under transmissibility I would read it as yes for the tick (therefore transmissible), and no for x (therefore not transmissible), which is exactly the opposite of what is meant there. Using it this way only leads to confusion. The colours alone are enough to indicate good or bad outcome. Hzh (talk) 23:48, 1 February 2021 (UTC)
- I agree, I initially created the table in the inverse - where ticks mean yes and crosses mean no. I understand that the red and green colours may have caused some confusion, so I have edited the table to remove the symbols altogether as you suggested. ArcMachaon (talk) 15:43, 2 February 2021 (UTC)
Cluster 5 not extinct?
German news n-tv reports that one dead in Bavaria from a strain with a Cluster 5-like mutation. It's just on their Covid ticker, but there should be some official or other usable RS; the source of the claim is not given more specifically than "Bavarian labs". 213.182.112.25 (talk) 22:41, 14 February 2021 (UTC)
- You say cluster 5–like mutation; does this mean only Y453F? Because in that case, it wouldn't be cluster 5 since there aren't necessarily the several other defining mutations. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 23:57, 14 February 2021 (UTC)
- A team from the SSI confirmed that Cluster 5 was not in circulation in the human population as of 1 February 2021 see here, section "Cluster 5 variant". I have added this to the section and removed the banner. Biscuit-in-Chief is likely correct in their assessment of the headline. If more information about the prevalence of specific mutations is found, feel free to add it to the "notable mutations" section. ArcMachaon (talk) 20:46, 18 February 2021 (UTC)
- The news item had no more details than "the strain found in Danisk mink farms" or similar. Thanks for the update, but it is not relevant to that case - the Feb 1 SSI paper is probably correct in Cluster 5 being extinct in Denmark (it would have been found since otherwise, because the Danes were actively looking for it), but the appearance in Bavaria was after Feb 1 (it was probably an infection that occurred locally at the end of January and no later than Feb 2, assuming the usual German testing/reporting workflow - the infected person was probably symptomatic as Germany does not routinely test people without symptoms, and add to this 2-4 days delay for reporting the original PCR test result, and another ~3 days before the sequencing results are reported), and how it would have gotten there is entirely mysterious. Given that Germany is rather lacklustre in its sequencing effort, I suspect that it's some Cluster-5-like convergent mutation, and that they confused it because only a few snippets are sequenced (Germany does apparently not distinguish between Lineage B.1.351 and B.1.525, or VOC-202012/01 and VOC-202102/02; tho both strains with additional E484K are reasonable to expect there given their spread elsewhere in Europe, none have been reported to date). I will try and find any follow-up to the Bavarian report, which is puzzling and if true kinda disturbing. 213.182.113.251 (talk) 13:08, 19 February 2021 (UTC)
- Ah yes, that was easy:
- https://www.swp.de/panorama/corona-mutation-bayern-neu-ulm-ansbach-daenemark-coronavirus-bayreuth-verbreitet-sich-b117-b1351-p1-zahlen-54700406.html
- https://www.stimme.de/suedwesten/nachrichten/pl/labor-entdeckt-daenische-corona-mutation-in-bayern;art19070,4443059
- https://www.merkur.de/welt/coronavirus-deutschland-bayern-mutation-nerz-cluster-5-neuulm-variante-tier-drosten-rki-90203580.html
- 10 cases with at least 1 fatality reported as of Feb 16; the first case reported Jan 27 already, meaning infection most likely occurred about Jan 10. 2 sequencing labs found it, one Austria (1 case), the other in Germany (the other cases), so it's likely to be genuine - though details are still lacking, it's just "Cluster 5" or "the Danish mink strain". Cases cluster in Neu-Ulm county, but at least one in Ansbach which is quite some way distant.
- The most consistent hypothesis is that someone on holiday or business travel picked it up in the very days before it was eradicated in Denmark. It's highly unlikely that more than 1, maybe 2 people imported the strain to Bavaria (probably Neu-Ulm area), given the distance involved; any spread beyond their household probably only happened during the Christmas/New Year week. That would result in the half-dozen or so chains of transmission known to be active at the end of January.
- I've put the links into the article (these are all local/regional daily newspapers, none of them "yellow press", and hence qualify as RS as long as no proper scientific source is available), but you may wanna tidy it up. I have copypasted it to the main Cluster 5 article too, but again, only quick-and-dirty. It should be OK for the meantime, but any tidying up (linking the sources; at least Münchner Merkur has an article) is also appreciated.)
- I hope there's a scientific study soon, because this is very puzzling (though not entirely implausible; it would still be awfully bad luck - literally "the one that got away") and worrying (Bavaria does a really bad job as regards containment; they neglect fighting domestic transmission in favour of closing external borders). At least they do sequence, though not yet (but hopefully soon) in an organized fashion. 213.182.113.251 (talk) 14:05, 19 February 2021 (UTC)
- @213.182.113.25: I have reverted your edits—the popular-media reports keep using to cluster 5 and "the Danish mink variant" interchangeably, which clearly shows they haven't understood it correctly: the SSI, as explained in this report, found five clusters of mink variants as of early November 2020; cluster 5 was just the concerning one. We should wait with reporting on this till there are reliable medical sources from subject-matter experts we can cite. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 15:21, 19 February 2021 (UTC)
- The news item had no more details than "the strain found in Danisk mink farms" or similar. Thanks for the update, but it is not relevant to that case - the Feb 1 SSI paper is probably correct in Cluster 5 being extinct in Denmark (it would have been found since otherwise, because the Danes were actively looking for it), but the appearance in Bavaria was after Feb 1 (it was probably an infection that occurred locally at the end of January and no later than Feb 2, assuming the usual German testing/reporting workflow - the infected person was probably symptomatic as Germany does not routinely test people without symptoms, and add to this 2-4 days delay for reporting the original PCR test result, and another ~3 days before the sequencing results are reported), and how it would have gotten there is entirely mysterious. Given that Germany is rather lacklustre in its sequencing effort, I suspect that it's some Cluster-5-like convergent mutation, and that they confused it because only a few snippets are sequenced (Germany does apparently not distinguish between Lineage B.1.351 and B.1.525, or VOC-202012/01 and VOC-202102/02; tho both strains with additional E484K are reasonable to expect there given their spread elsewhere in Europe, none have been reported to date). I will try and find any follow-up to the Bavarian report, which is puzzling and if true kinda disturbing. 213.182.113.251 (talk) 13:08, 19 February 2021 (UTC)
- The previously linked swp.de source says: "Am Mittwochabend ließ das Landratsamt Neu-Ulm jedoch vermelden, dass ihnen Informationen über einen gesicherten Nachweis der dänischen Variante in Proben aus Bayern bislang nicht vorliegen" (translated: "Wednesday the Neu-Ulm District office said that they did not have information about reliable evidence showing the Danish variant in Bavaria"). Additionally, the sources keep on randomly going between "Cluster 5", "Danish variant/Danish mink variant" and "mink variant". These are not the same thing. As mentioned by Biscuit-in-Chief, cluster 5 is a very specific mink variant (hence its name, "5"), which only ever was found in 12 people in/near one village several months ago. A number of other mink-related variants do exist and they have been involved in more cases in a wider region, but they're not known to be particularly problematic; their behavior is similar to "ordinary" COVID-19. All mink-related in Denmark have, collectively, been referred to as the "Danish (mink-related) variants"; all these are now extinct in the country with no recent detections. Then there are mink-related variants without further specification, which have been discovered in several countries with outbreak in mink farms. They're generally defined by the Y453F mutation in combination with others, and part of several different clades (i.e., they emerged independently of each other). However, the problem is that the Y453F mutation, while usually linked to mink, also can occur under other circumstances, as shown by its developent in an immunocompromised, long covid patient (Δ69-70 itself is quite common, having been detected in at least six independent clades, with first detection more than a year ago in Thailand and Germany). I've considered updating/correcting the Cluster 5 article and will probably do it one of the next days, including its "Implications for human health" section which confuses cluster 5 (=12 cases) and "Danish (mink-related) variants" (=more cases); this is also described in the recently published review. However, until we get clear information about the German cases, it is impossible to say if or where they belong in this, but I'd be very surprised if they're real cluster 5. They're in all probability "Danish (mink-related) variant" or some other variant with a Y453F mutation. I've just checked GISAID and Nextstrain and they have no cluster 5/Danish (mink-related) variant/Y453F mutation from Germany. Finally, I am very puzzled by the claim in the most recent German article that these have been little studied; there have been a good handful of scientific articles about them. RN1970 (talk) 00:24, 20 February 2021 (UTC)
Seven U.S. variants
Emergence in late 2020 of multiple lineages of SARS-CoV-2 Spike protein variants affecting amino acid position 677 (not peer reviewed as of now) This study is showing up in a number of mainstream media reports. Mapsax (talk) 22:59, 15 February 2021 (UTC)
- yes Ive seen it as well--Ozzie10aaaa (talk) 20:56, 16 February 2021 (UTC)
Vandalism / Spamming
ZRS2012 seems to be repeatedly vandalizing this page, and has done similar things to other covid related pages. Is there any recourse? TimeEngineer (talk) 00:50, 18 February 2021 (UTC)
- Ive got this article on my watchlist--Ozzie10aaaa (talk) 02:27, 18 February 2021 (UTC)
- Surely the user:ZRS2012 must be in violation, saying he/she is the same as recently blocked user:Theusernameistaken, who indeed as been blocked indefinitely. 178.155.171.181 (talk) 19:35, 18 February 2021 (UTC)
- On this basis, how many warnings are required on the user:ZRS2012 talk page before blocking further edits from them can be requested? This seems a somewhat aggravated case -see WP:AIV and the warning templates - I see that Biscuit-in-Chief has already posted a 'vandalism3' ("please stop") notice at User talk:ZRS2012 -Hope this helps, Yadsalohcin (talk) 23:28, 18 February 2021 (UTC)
- Yeah, we can probably request that now. I posted vandalism3 before their most recent vandalism. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 23:42, 18 February 2021 (UTC)
- Actually, we most definitely can, per WP:EVADE. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 23:44, 18 February 2021 (UTC)
- Yeah, we can probably request that now. I posted vandalism3 before their most recent vandalism. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 23:42, 18 February 2021 (UTC)
- On this basis, how many warnings are required on the user:ZRS2012 talk page before blocking further edits from them can be requested? This seems a somewhat aggravated case -see WP:AIV and the warning templates - I see that Biscuit-in-Chief has already posted a 'vandalism3' ("please stop") notice at User talk:ZRS2012 -Hope this helps, Yadsalohcin (talk) 23:28, 18 February 2021 (UTC)
- Surely the user:ZRS2012 must be in violation, saying he/she is the same as recently blocked user:Theusernameistaken, who indeed as been blocked indefinitely. 178.155.171.181 (talk) 19:35, 18 February 2021 (UTC)
@Yadsalohcin and TimeEngineer: I've reported them at Wikipedia:Administrators'_noticeboard/Incidents#ZRS2012_evading_block_and_doing_disruptive_edits. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 23:55, 18 February 2021 (UTC)
- They're now blocked indefinitely. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 00:17, 19 February 2021 (UTC)
@Biscuit-in-Chief: They seem to be back as "GaatWild". Same sentence structure in the comments and similar revisions TimeEngineer (talk) 21:06, 23 February 2021 (UTC)
- @TimeEngineer: WP:SPI. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 21:26, 23 February 2021 (UTC)
- @Biscuit-in-Chief: done here link TimeEngineer (talk) 14:35, 25 February 2021 (UTC)
@Ifnord:, @Biscuit-in-Chief: and @TimeEngineer: we seem to have a resumption of this disruptive vandalism, previously by User:ZRS2012 and User talk:GaatWild, this time from User:DORKKLOK. I see Ifnord has placed a warning template on their user page (thanks) and I have now alerted the admin previously involved (LuK3 (Talk)). Yadsalohcin (talk) 22:00, 5 March 2021 (UTC)
Spanish variant
In October 2020, a Spanish variant of COVID-19 was discovered, and known as 20A.EU1. This is what caused the second wave of infections in the UK and other European countries following the "travel corridors" idea. The article stated that the discovery had not been peer-reviewed so this is probably why this wasn't given a mention here... though I wonder if it could this time now that I've brought it up. What does everyone think? Bryn89 (talk) 18:12, 21 February 2021 (UTC)
- This is also known as B.1.177 and appears in the table as a clade. Though it contains the A222V mutation, it doesn't seem to affect transmissibility - it was just the strain that holidaymakers happened to bring back from Spain at the end of summer (see Sci Am and BBC Future. If you can find review articles discussing it then a brief section could be added. One says "Prior to B.1.1.7, B.1.177 (also called “20A.EU1”) was the dominant variant in the UK, although it is not believed to have arisen due to increased transmissibility. Since the earliest sequenced sample of B.1.1.7 (September 20, 2020), it started to displace B.1.177 and other variants in the UK."[2] Fences&Windows 21:42, 25 February 2021 (UTC)
- I do think that this variant received enough coverage in media and medical sources to fulfil the notability criteria. It is also notable because it became a main lineage and is still the dominant strain in parts of Europe. I also note it is mentioned on the SARS-CoV-2 main page but not here:
In July 2020, scientists reported that a more infectious SARS-CoV-2 variant with spike protein variant G614 has replaced D614 as the dominant form in the pandemic. In October 2020 scientists reported in a preprint that a variant, 20A.EU1, was first observed in Spain in early summer and has become the most frequent variant in multiple European countries. They also illustrate the emergence and spread of other frequent clusters of sequences using Nextstrain.
- Overall it would probably be a good WP:BOLD addition to this page. ArcMachaon (talk) 14:49, 26 February 2021 (UTC)
Use of {{risk}} in summary table
There seems to be some confusion with the colours in the summary table which seem subjective and not based upon a system or any reliable source. I would suggest that there are two options to fix this problem: return to the red-yellow-green (bad-pending-good) system previously used, or create a system a place it in comment form above the table for editors to see. For example:
<!--
Table Colours for clinical changes:
Risk should be "unknown" if risk is under investigation
Risk should be "low" if risk has been disproven by reliable investigation
Risk should be "medium" when "potentially", "likely", "indications of" etc. (NB. Reliable medical source still required)
Risk should be "high" if confirmed
Risk should be "very high" if critical and confirmed
Table Colours for spread:
Risk should be "very low" if thought extinct
Risk should be "low" if contained and localised
Risk should be "medium" if endemic to a specific region
Risk should be "high" if global spread
Risk should be "very high" if variant becomes a major lineage per reliable medical sources
-->
as one potential system. What are people's thoughts? ArcMachaon (talk) 23:53, 26 February 2021 (UTC)
- It is sensible to define better criteria. I'd seek some inspiration on the example risk matrix, though every risk matrix is different. For clinical changes, I'd consider risk as a combination of probability and harm level. When the harm level refers to an increase in expected deaths, I'd suggest basing the choices on the "critical" harm level with some adjustments, like this:
- "unknown" if risk is under investigation
- "verylow" if probability is described as rare or if risk has been disproven to a degree by reliable investigation or even "eliminated" if investigators are very confident of their results
- "low" if probability is described as unlikely
- "medium" if probability is described as possible or potential or as having some indications
- "high" if probability is described as likely
- "veryhigh" if probability is described as certain
- When risk refers to the harm level of disease leading lost work days but no deaths and no permanent disability, then I'd follow the "marginal" harm level. It is important to evaluate both probability and harm properly. For example, P.1. is currently described as "10–80% more lethal", but the link to CADDE says "can be 10–80% more lethal", meaning "potentially 10–80% more lethal". --Fernando Trebien (talk) 00:39, 8 March 2021 (UTC)
- Perhaps risk assessment framework from Public Health England can help refine the current criteria.[1] For example:
Changes Efficacy or neutralizing antibody activity Transmissibility Hospitalization / Mortality After natural infection From vaccination Monoclonal antibodies No transmission Decrease confirmed Evidence of no change Evidence of no change Evidence of no change Limited to clusters Similar to previous variants Data suggesting a difference Data suggesting a difference Data suggesting a difference Similar to previous variants Increase confirmed but limited to risk groups Experimental evidence of evasion Experimental evidence of evasion Experimental evidence of decreased efficacy Increase confirmed Increase confirmed for most groups Evidence of frequent reinfection Evidence of decreased efficacy Evidence of decreased efficacy
Fin-796H variant (B.1.1.318 Lineage)
Newly discovered Fin-796H variant is likely to have some historical significance as it includes a mutation which makes it harder to detect with some PCR-tests, bears similarities to both British and South African variants (E484K mutation included) and appears to be from a distinct lineage that has gone unnoticed for some time. It is unlikely to have originated in Finland where it was first discovered, as the local rate of infection is relatively low and this variant is said to contain at least 15 distinct mutations. Not much seems to be known yet about how widespread, contagious or vaccine-resistant it is, however it was recently sequenced and had it's sequence sent to GISAID. I'll be leaving inclusion decisions and wording choices up to someone more experienced... Sources (in English): https://yle.fi/uutiset/osasto/news/new_coronavirus_variant_discovered_in_finland/11796958 https://www.coronaheadsup.com/coronavirus/second-case-of-variant-fin-796h-confirmed-in-finland-four-more-fin796h-infections-likely/ Nuihc88 (talk) 07:21, 7 March 2021 (UTC)
- I think it was discovered some almost a month ago, so it isn't quite recent, but agree that it seems notable. What's actually recent is a French variant discovered a week ago which also fails on all three PCR gene targets. Ain92 (talk) 12:28, 24 March 2021 (UTC)
Looks like this variant is part of the B.1.1.318 lineage and most likely originated in Nigeria, but all the news reporting i could find about it seems shoddy and lacks primary sources. Nuihc88 (talk) 20:08, 19 April 2021 (UTC)
Indian "double" mutant
It looks like another variant has been discovered in India: https://www.bbc.com/news/world-asia-india-56507988 — Preceding unsigned comment added by 2A01:799:B22:4F00:CD72:8884:932F:220 (talk) 19:26, 24 March 2021 (UTC)
- India reports 'double mutant' coronavirus variant as daily deaths reach year's high CBCyes interesting--Ozzie10aaaa (talk) 19:33, 24 March 2021 (UTC)
Proposal for shorthand redirect page...?
Hello, I'm sorry. I was curious if maybe a few pages for shorthand phrases like "COVID-19 VOC" could be created to redirect to this article since it seems natural to me that people would use such keywords to get to this page quickly and more often than typing out "variants of SARS-CoV-2" or something. I'd make the redirect of it myself but I actually don't know if I'm allowed to do so (especially as a guest user) or if I'd have to make a request to have it done instead. Thank you. 65.92.88.128 (talk) 22:25, 24 March 2021 (UTC)
- COVID variant already exists, although I suppose we could create "COVID-19 VOC" as well. JackFromReedsburg (talk | contribs) 22:31, 24 March 2021 (UTC)
New variant A.VOI.V2 from Tanzania
South African scientists identified the most genetically divergent variant so far, see [3]. Ain92 (talk) 11:01, 27 March 2021 (UTC)
- "A novel variant of interest of SARS-CoV-2 with multiple spike mutations detected through travel surveillance in Africa". www.krisp.org.za. Retrieved 30 April 2021.--Ozzie10aaaa (talk) 22:39, 30 April 2021 (UTC)
New Variant ‘double mutant’ strain named B.1.617
Hi, I'm new to Wiki but I just came across the new variant B.1.617 and see it's not added. I don't know what I'm doing so I just though I would mention it. Thanks to everyone and their hard work here. — Preceding unsigned comment added by RegCliff (talk • contribs) 23:00, 8 April 2021 (UTC)
- thank you for link--Ozzie10aaaa (talk) 23:04, 8 April 2021 (UTC)
Legend for Table Colors
Hello there, I see the table colors are unintuitive, it's not clear what the yellow and red and green are denoting. I would love someone could add a legend or otherwise visual assistant to explain the color coding Buumix (talk) Boring 13:45, 19 April 2021 (UTC)
- Done. --Fernando Trebien (talk) 02:39, 23 May 2021 (UTC)
New variant B.1.618
Hi, can anyone well versed with the variant's notability, add about new variant B.1.618[2][3][4] that been supposed to be originated in India and has been found in US, Switzerland, Singapore and Finland as per reports.[5] Thank you. Run n Fly (talk) 14:38, 21 April 2021 (UTC)
References
- ^ "SARS-CoV-2 variants: risk assessment framework" (PDF). GOV.UK. Government Digital Service. Public Health England. 2021-05-22. GOV-8426.
- ^ Koshy, Jacob (21 April 2021). "New coronavirus lineage found in West Bengal". The Hindu.
- ^ Dasgupta, Binayak; Dutt, Anonna (21 April 2021). "New B.1.618 variants now among most sequenced". Hindustan Times.
- ^ Basu, Mohana (20 April 2021). "New Covid lineage B.1.618 identified from Bengal, 2nd in India after 'double mutant' virus". ThePrint.
- ^ Sharma, Milan (21 April 2021). "New immune escape coronavirus variant found in West Bengal, say experts". India Today.
New York Variant - Lineage B.1.526
Hello à tous,
I would like to inform you that a page is ready to be linked about the New York Variant: Lineage B.1.526. As I don't know how to insert it in this article, I have preferred to write a comment in the talk page, so that you guys who know more about the subject, would see how to deal with it, without adding in the See also section.
Thanks for all the work you do for the vulgarisation of this damned virus/disease/pandemic. --Anas1712 (talk) 23:58, 21 April 2021 (UTC)
 Partly done and anyone is welcomed to expand. Run n Fly (talk) 07:20, 22 April 2021 (UTC)
Partly done and anyone is welcomed to expand. Run n Fly (talk) 07:20, 22 April 2021 (UTC)
Thank you to editors
I thank all of the reasonable and hard-working editors who have been meticulously updating this extremely important article, which is well-structured and well-written, and its related companion articles. Acwilson9 (talk) 08:08, 24 April 2021 (UTC)
P1 > Assessment source
- Freitas, André Ricardo Ribas; Lemos, Daniele Rocha Queiróz; Beckedorff, Otto Albuquerque; Cavalcanti, Luciano Pamplona de Góes; Siqueira, Andre M.; Mello, Regiane Cristina Santos de; Barros, Eliana N. C. (2021-04-19). "The increase in the risk of severity and fatality rate of covid-19 in southern Brazil after the emergence of the Variant of Concern (VOC) SARS-CoV-2 P.1 was greater among young adults without pre-existing risk conditions". medRxiv: 2021.04.13.21255281. doi:10.1101/2021.04.13.21255281.
New variant B1616
I have seen news articles about B1616 and that it can evade PCR tests. Shall we add em in? Hanami-Sakura (talk) 07:24, 12 May 2021 (UTC)
- Do you mean B.1.617? 'Evading' PCR sounds weird. Can you provide links. JuanTamad (talk) 07:48, 12 May 2021 (UTC)
https://www.medrxiv.org/content/10.1101/2021.05.05.21256690v1
https://www.ouest-france.fr/sante/virus/coronavirus/covid-19-la-direction-generale-de-la-sante-alerte-sur-un-nouveau-variant-detecte-en-bretagne-7187918 Unxed (talk) 13:33, 19 May 2021 (UTC)
Dates in table
We have B.1.1.7 listed as first identified in February 2020, but it wasn't detected until December and was later found to have first been sequenced in September. The cited source, https://cov-lineages.org/lineages/lineage_B.1.1.7.html, does say February but it is clearly wrong: https://asm.org/Articles/2021/January/B-1-1-7-What-We-Know-About-the-Novel-SARS-CoV-2-Va ; https://www.sciencemag.org/news/2020/12/mutant-coronavirus-united-kingdom-sets-alarms-its-importance-remains-unclear. Do we need to stop using cov-lineages.org? Fences&Windows 11:31, 12 May 2021 (UTC)
- thank you for posting--Ozzie10aaaa (talk) 01:36, 24 May 2021 (UTC)
- February 2020 is the the date given by outbreak.info as well for any detection in the world. September 2020 is when it was first detected in the UK. --Fernando Trebien (talk) 18:03, 6 June 2021 (UTC)
- But I asked about this in the issue tracker of cov-lineages and they said that the lineages pages are not quality controlled and that there is a lot of data with incorrect labels. In cov-lineages, the best source for this information is the global report. It seems that outbreak.info also has no quality control either and it uses the same data source (GISAID). So it is better to rely on reviewed information provided by trusted sources such as the WHO, PHE, CDC, ECDC, ACI. --Fernando Trebien (talk) 11:29, 14 June 2021 (UTC)
Why do we still retain B.1.1.207 in the overview table?
Looks not so notable actually. Ain92 (talk) 19:46, 20 May 2021 (UTC)
- Agreed, and removed. --Fernando Trebien (talk) 01:48, 23 May 2021 (UTC)
Strains missing from overview table
The WHO "variants of interest"[4] now called Zeta, Theta, and Iota are missing from the Overview table. Given the WHO is worried about them, seems useful to include them. -- Beland (talk) 18:19, 31 May 2021 (UTC)
WHO names variants based on Greek alphabet?
Seems WHO has reported (31 May 2021) a new system of identifying Covid-19 variants - based on the Greek alphabet[1][2] - examples: the "UK variant" (B.1.1.7) is "Alpha"; the "South African variant" (B.1.351) is "Beta" - and so forth - perhaps this new system should be added to the main article? - in any case - Stay Safe and Healthy !! - Drbogdan (talk) 01:20, 1 June 2021 (UTC)
 Done - Brief followup - seems this has already been done - Stay Safe and Healthy !! - Drbogdan (talk) 02:00, 1 June 2021 (UTC)
Done - Brief followup - seems this has already been done - Stay Safe and Healthy !! - Drbogdan (talk) 02:00, 1 June 2021 (UTC)
References
- ^ Staff (31 May 2021). "SARS-CoV-2 Variants of Concern and Variants of Interest, updated 31 May 2021". World Health Organization. Retrieved 31 May 2021.
- ^ Howard, Jacqueline (31 May 2021). "WHO's new naming system for coronavirus variants uses Greek alphabet". CNN News. Retrieved 31 May 2021.
News (06/02/2021) worth considering?
Recent news (06/02/2021)[1] worth considering for the main article? - iac - Stay Safe and Healthy !! - Drbogdan (talk) 00:25, 3 June 2021 (UTC)
References
- ^ Speights, Keith (2 June 2021). "This Ominous Warning From Moderna Could Shake Up the COVID Vaccine Market - The pandemic isn't over yet -- even in the U.S." The Motley Fool. Retrieved 2 June 2021.
Pangolin origin information needs updating
This Though the emergence of SARS-CoV-2 may have resulted from recombination events between a bat SARS-like coronavirus and a pangolin coronavirus (through cross-species transmission)
needs update because more recent research downplays the possible role of pangolins for SARS-CoV-2 evolutionary history. See Severe Acute Respiratory Syndrome Coronavirus 2 for more current sources and up-to-date information. Forich (talk) 21:18, 4 June 2021 (UTC)
A Nature article by the Nextstrain team about a non-notable, yet quite interesting variant 20E(EU1)/B.1.177
[5]. I have no idea in what Wikipedia article it is best applicable, but found an overview Twitter thread to be interesting, take a look: https://twitter.com/firefoxx66/status/1401833676317593600 Ain92 (talk) 11:45, 7 June 2021 (UTC)
Date of sequence of B.1.1.7
The article states that Lineage B.1.1.7 was sequenced in October 2020, sourced on a BBC article. I have this source[1] that says it was in late september. Which one is more accurate? Forich (talk) 20:05, 11 June 2021 (UTC)
- At the moment, your source is more accurate and the date given (20 September 2020) is exactly the same as given in the UK by cov-lineages and outbreak.info. The news piece referenced in the article was probably correct at the time it was published (December 2020) because it often takes time for new data to be integrated into the main databases. Note that this is not the oldest sequence in the world, which is February 2020, as reported in the Overview section. --Fernando Trebien (talk) 12:15, 13 June 2021 (UTC)
- Thanks, I believe it is updated now. Forich (talk) 20:43, 15 June 2021 (UTC)
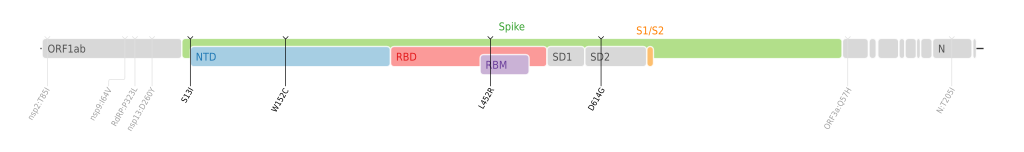
Genome maps
We (the Stanford HIVDB team) created these figures for different SARS-CoV-2 variants since this year, and we just released it under CC-BY-SA 4.0. I have uploaded the top four WHO named variants to Commons. Does anyone else other than me also think those figures are useful to be included in the variant articles? I'm afraid it is kind of interest conflict if I add those figures to the articles.
Best. --PhiLiP (talk) 19:06, 21 June 2021 (UTC)
- @PhiLiP: They seem pretty neat and useful. —hueman1 (talk • contributions) 08:36, 26 June 2021 (UTC)
- @HueMan1: Thank you very much! I have added more. Can you also help me to add them to the corresponding articles for me to avoiding conflicts of interest? Thanks!
- Best. --PhiLiP (talk) 17:35, 30 June 2021 (UTC)
- ^ Washington, Nicole L.; Gangavarapu, Karthik; Zeller, Mark; Bolze, Alexandre; Cirulli, Elizabeth T.; Schiabor Barrett, Kelly M.; et al. (May 2021). "Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States". Cell. 184 (10): 2587–2594.e7. doi:10.1016/j.cell.2021.03.052.
- List-Class Molecular Biology articles
- Low-importance Molecular Biology articles
- All WikiProject Molecular Biology pages
- List-Class COVID-19 articles
- High-importance COVID-19 articles
- WikiProject COVID-19 articles
- List-Class medicine articles
- Low-importance medicine articles
- List-Class emergency medicine and EMS articles
- Mid-importance emergency medicine and EMS articles
- Emergency medicine and EMS task force articles
- List-Class society and medicine articles
- Mid-importance society and medicine articles
- Society and medicine task force articles
- List-Class pulmonology articles
- High-importance pulmonology articles
- Pulmonology task force articles
- All WikiProject Medicine pages
- List-Class virus articles
- Mid-importance virus articles
- WikiProject Viruses articles