Topoisomerase
| DNA Topoisomerase, ATP-independent (type I) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 5.6.2.1 | ||||||||
| CAS no. | 80449-01-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
| DNA Topoisomerase, ATP-dependent (type II) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 5.6.2.2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
DNA topoisomerases (or topoisomerases) are enzymes that catalyze changes in the topological state of DNA, interconverting relaxed and supercoiled forms, linked (catenated) and unlinked species, and knotted and unknotted DNA.[1][2] Topological issues in DNA arise due to the intertwined nature of its double-helical structure, which, for example, can lead to overwinding of the DNA duplex during DNA replication and transcription. If left unchanged, this torsion would eventually stop the DNA or RNA polymerases involved in these processes from continuing along the DNA helix. A second topological challenge results from the linking or tangling of DNA during replication. Left unresolved, links between replicated DNA will impede cell division. The DNA topoisomerases prevent and correct these types of topological problems. They do this by binding to DNA and cutting the sugar-phosphate backbone of either one (type I topoisomerases) or both (type II topoisomerases) of the DNA strands. This transient break allows the DNA to be untangled or unwound, and, at the end of these processes, the DNA backbone is resealed. Since the overall chemical composition and connectivity of the DNA do not change, the DNA substrate and product are chemical isomers, differing only in their topology.
Discovery
The first DNA topoisomerase was discovered in bacteria by James Wang in 1971 and was initially named ω (omega) protein;[3] it is now called Escherichia coli (E. coli) topoisomerase I (topo I) and is a representative of the type IA family of enzymes. Subsequently, a similar activity was found in eukaryotic cells (rat liver) by James Champoux and Renato Dulbecco;[4] the enzyme responsible, eukaryotic topo I, has a distinct mechanism and is representative of the type IB family. The first type II topoisomerase to be discovered was DNA gyrase from bacteria, by Martin Gellert and coworkers in 1976 [5]0, and also characterized by Nicholas Cozzarelli and co-workers.[6] DNA gyrase catalyzes the introduction of negative supercoils into DNA and is the only type II enzyme to do this, all the others catalyze DNA relaxation. Type II enzymes are mechanistically distinct from type I in being ATP-dependent and transiently cleaving both DNA strands rather than just one. Type II topoisomerases were subsequently identified from bacterial viruses and eukaryotes.[7][8][9] Topo EC-codes are as follows: ATP-independent (type I), EC 5.6.2.1; ATP-dependent (type II): EC 5.6.2.2. The exception among the type I topoisomerases, reverse gyrase, which contains a helicase domain (EC 3.6.4.12) and introduces positive supercoiling in an ATP-dependent manner. Therefore it is the sole type I topoisomerase classified as EC 5.6.2.2 (Table 1).
DNA Topology
DNA Topology (topological problems in cellular DNA). The double-helical structure of DNA involves the intertwining of the two polynucleotide strands around each other, which potentially gives rise to topological problems. DNA topology refers to the crossing of the two DNA strands that alters the twist of the double helix and gives rise to tertiary conformations of DNA, such as supercoils, knots and catenanes.[10] Potential topological issues associated with the double-helical structure of DNA were recognized soon after its structure was first elucidated in 1953 by James Watson, Francis Crick and Rosalind Franklin[11][12][13] and developed further by the work of Max Delbruck and John Cairns.[14][15] Closed-circular double-stranded DNA can be described by 3 parameters: Linking number (Lk), Twist (Tw) and Writhe (Wr) (Fig. 1). Where Lk refers to the number of times the two strands are linked, Tw refers to the number of helical turns in the DNA, measured relative to the helical axis, and Wr quantifies the coiling of the path of the DNA helix in space and is often equated with ‘supercoiling’. The 3 parameters are related as follows: Lk = Tw +Wr. This is a mathematical identity originally obtained by Călugăreanu in 1959 (16) and is referred to as the Călugăreanu, or Călugăreanu–White–Fuller, theorem (17,18). Lk cannot be altered without breaking one or both strands of the helix; Tw and Wr are interconvertible and depend upon the solution conditions. Supercoiling is a vernacular term for DNA with a non-zero linking difference, more correctly referred to as specific linking difference (σ = ΔLk/Lk0, where Lk0 is the mean linking number of the relaxed DNA circle). DNA is said to be positively supercoiled if Lk of it is higher than Lko for the relaxed state (Lk-Lko = ΔLk, ΔLk>0); that means that Tw and/or Wr are increased relative to the relaxed molecule. Conversely, DNA is negatively supercoiled if Lk of the molecule is lower than the Lko (ΔLk<0).
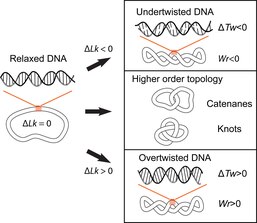
The overall function of DNA topoisomerase is to manage the topological state of the DNA in the cell. There are two types or families of this enzyme; type I family and type II family. Type I family passes one strand of the DNA through a break in the opposing strand. In other words, DNA topoisomerase type I enzyme cleaves only one strand of DNA. Type II family passes a region of duplex (two strands) from the same molecule or a different molecule through a double stranded gap. To summarize, type II cleaves both strands of DNA, that results in a double-stranded break. Topoisomerases can either relieve negative supercoils, both positive and negative supercoils, or induce positive and negative supercoiling in DNA. The enzymes also can promote catenation (when two single circular DNA strands are linked together after replication) and decatenation (the separation of two linked, closed, circular chromosomes), and can also relieve entanglement of linear chromosomes.[16]
The double-helical configuration of DNA strands makes them difficult to separate, which is required by helicase enzymes if other enzymes are to transcribe the sequences that encode proteins, or if chromosomes are to be replicated. In circular DNA, in which double-helical DNA is bent around and joined in a circle, the two strands are topologically linked, or knotted. Otherwise identical loops of DNA, having different numbers of twists, are topoisomers, and cannot be interconverted without the breaking of DNA strands. Topoisomerases catalyze and guide the unknotting or unlinking of DNA[17] by creating transient breaks in the DNA using a conserved tyrosine as the catalytic residue.[18]
The insertion of (viral) DNA into chromosomes and other forms of recombination can also require the action of topoisomerases.
Topologically linked circular molecules, aka catenanes, adopt a positive supercoiled form during the process of replication of circular plasmids. The unlinking of catenanes is performed by type IIA topoisomerases, which were recently found to be more efficient unlinking positive supercoiled DNA.The conformational properties of negative vs. positive supercoiled catenanes affects their features in respect to their corresponding enzymatic reaction catalyzed by topoisomerases. Experiments have demonstrated that positive supercoiled DNA provides a sharp DNA bend in the first bound DNA segment, which allows the topoisomerase to bind successfully and therefore carry on its enzymatic reaction to the following segment in a specific inside-to-outside matter. On the other hand, negative supercoiled DNA does not provide such bend and the access of the enzyme to the first segment is nearly impossible, therefore inhibiting unlinking.[19]
Topoisomerase is also found in the mitochondria of cells.[20] The mitochondria generate ATP as well as playing a role in programmed cell death and aging.[21] The mitochondrial DNA of animal cells is a circular, double-stranded DNA that requires the activity of topoisomerase to be replicated. The classes of topoisomerase found in the mitochondria are I, IIβ, IIIα.[20]
Yeast
Yeast cells are known to use three topoisomerases: Topoisomerase I, from the IB subfamily, is required for growth. It provides the replication fork with the ability to move forward, as well as removes positive and negative supercoils associated with transcription. Topoisomerase II from the IIA subfamily, is needed for decatenation of linked chromosomes and preparation for segregation during mitosis. Topoisomerase II cannot induce negative supercoils, but can relax both positive and negative supercoils like topoisomerase I, and can replace topoisomerase I if absent. Topoisomerase III from the IA family is used for cell growth. Without topoisomerase III, recombination rates in mitosis and meiosis can increase, which slows growth in cells. In S. pombe cells, III is used to sustain cell division.
Higher eukaryotes
Higher eukaryotic organisms are more complex organisms and typically require more complex cellular machinery. These organisms generally have a topoisomerase I, two type IIA topoisomerases, and two type III enzymes. Topoisomerase I helps with replication fork movement and relaxes supercoils associated with transcription. It is also used for relaxing solenoidal supercoils that form when chromosomes condense in preparation for mitosis. The two type IIA topoisomerases, IIα and IIβ, are used to unlink intertwined daughter duplexes, as well as assist in cell division and suppression of recombination, respectively. Type IIIα and IIIβ are thought to work in embryogenesis and interact with helicases, respectively.
Eubacteria
E. coli contains four DNA topoisomerases: two type IA enzymes (I and III), and two type IIA (DNA gyrase and topoisomerase IV). DNA topoisomerase III and IV have similar functions. Topoisomerase III is incapable of relaxing positive supercoils, but it works to support replication fork movement on plasmid DNA in vitro. It can decatenate the winding that is happening behind the replication fork by focusing on nicks in the DNA. Topoisomerase IV is the most effective decatenating enzyme in E. coli. It also relaxes negative supercoils. DNA gyrase uses the hydrolysis of ATP to generate negative supercoiling in bacterial chromosomes. It relaxes positive supercoils ahead of the replication fork and acts in chromosome condensing. Finally, topoisomerase I helps with generating some negative supercoiling along with topoisomerase IV and DNA gyrase.
Archaea
There is limited knowledge about the archaeal genome sequences. Therefore, there is also limited knowledge about the topoisomerase enzymes. They do contain a reverse gyrase, a type IA topoisomerase, and topoisomerase VI. The functions of the topoisomerases in archaea are comparable to the enzymes in eubacteria. The only noteworthy difference is that topoisomerase VI in archaea is responsible for decatenation of DNA replication intermediates, and it relaxes both positive and negative supercoils.
Bacteriophage
The bacteriophage (phage) T4 gyrase (type II topoismerase) is a multisubunit protein consisting of the products of genes 39, 52 and probably 60. It catalyses the relaxation of negatively or positively superhelical DNA and is employed in phage DNA replication during infection of the E. coli bacterial host. Since the host E. coli DNA gyrase can partially compensate for the loss of the phage T4 gene products, mutants defective in either genes 39, 52 or 60 do not completely abolish phage DNA replication, but rather delay its initiation.[22] Mutants defective in genes 39, 52 or 60 show increased genetic recombination as well as increased base-substitution and deletion mutation suggesting that the host compensated DNA synthesis is less accurate than that directed by wild-type phage.[23] Mutants defective in genes 39, 52 and 60 also have reduced ability to carry out multiplicity reactivation, a form of recombinational repair that can deal with different types of DNA damage.[24]
DNA topology
DNA topology is the tertiary conformations of DNA, such as supercoiling, knotting, and catenation. Topology of DNA can be disrupted by most metabolic processes: RNA polymerase can cause positive supercoils by over-winding the DNA in front of the enzyme, and can also cause negative supercoils by under-winding the DNA behind the enzyme. DNA polymerase has the same effect in DNA replication. Positive and negative supercoiling balance out the entire global topology of the DNA, so overall, the topology remains the same. However, as the DNA replication or transcription fork moves forward and positive supercoiling increases, the DNA strands wrap tighter and tighter around each other, making it more difficult for the polymerase to move forward. It is important for the local topology of DNA ahead of and behind the polymerase to be relieved so that replication and cell division can proceed. This is what DNA topoisomerases are used for.[25]
Topological problems
There are three main types of problems:
Outside of the essential processes of replication or transcription, DNA must be kept as compact as possible, and these three states help this cause. However, when transcription or replication occurs, DNA must be free, and these states seriously hinder the processes. In addition, during replication, the newly replicated duplex of DNA and the original duplex of DNA become intertwined and must be completely separated in order to ensure genomic integrity as a cell divides. As a transcription bubble proceeds, DNA ahead of the transcription fork becomes overwound, or positively supercoiled, while DNA behind the transcription bubble becomes underwound, or negatively supercoiled. As replication occurs, DNA ahead of the replication bubble becomes positively supercoiled, while DNA behind the replication fork becomes entangled forming precatenanes. One of the most essential topological problems occurs at the very end of replication, when daughter chromosomes must be fully disentangled before mitosis occurs. Topoisomerase IIA plays an essential role in resolving these topological problems.
Clinical significance
Many drugs operate through interference with the topoisomerases.[26]
The broad-spectrum fluoroquinolone antibiotics act by disrupting the function of bacterial type II topoisomerases. These small molecule inhibitors act as efficient anti-bacterial agents by hijacking the natural ability of topoisomerase to create breaks in chromosomal DNA.
Some chemotherapy drugs called topoisomerase inhibitors work by interfering with mammalian-type eukaryotic topoisomerases in cancer cells. This induces breaks in the DNA that ultimately lead to programmed cell death (apoptosis). This DNA-damaging effect, outside of its potential curative properties, may lead to secondary neoplasms in the patient.[citation needed]
Topoisomerase I is the antigen recognized by Anti Scl-70 antibodies in scleroderma.
Classes
Topoisomerases can fix these topological problems and are separated into two types depending on the number of strands cut in one round of action.[27] Both these classes of enzyme utilize a conserved tyrosine; however, these enzymes are structurally and mechanistically different. For a video of this process click here.
- A type I topoisomerase cuts one strand of a DNA double helix, relaxation occurs, and then the cut strand is re-ligated. Cutting one strand allows the part of the molecule on one side of the cut to rotate around the uncut strand, thereby reducing stress from too much or too little twist in the helix. Such stress is introduced when the DNA strand is "supercoiled" or uncoiled to or from higher orders of coiling. Type I topoisomerases that do not require ATP for hydrolysis are subdivided into three subclasses:
- Type IA topoisomerases, which share many structural and mechanistic features with the type II topoisomerases. Examples of type IA topoisomerases include prokaryotic topoisomerase I and III, eukaryotic topoisomerase IIIα and topoisomerase IIIβ and reverse gyrase. Like type II topoisomerases, type IA topoisomerases form a covalent intermediate with the 5' end of DNA.
- Type IB topoisomerases, which utilize a controlled rotary mechanism. Examples of Type IB topoisomerases include Eukaryotic and eukaryal viral topoisomerase I. In the past, type IB topoisomerases were referred to as eukaryotic topoisomerase I, but IB topoisomerases are present in all three domains of life. Type IB topoisomerases form a covalent intermediate with the 3' end of DNA.
- Type IC topoisomerase (also called topoisomerase V) has been identified.[28] While it is structurally unique from type IA and IB topoisomerases, It shares a similar mechanism with type IB topoisomerase.
- A type II topoisomerase cuts both strands of one DNA double helix, passes another unbroken DNA helix through it, and then re-ligates the cut strands. Type II topoisomerases utilize ATP hydrolysis and are subdivided into two subclasses which possess similar structure and mechanisms:
- Type IIA topoisomerases which include eukaryotic and eukaryal viral topoisomerase IIα and topoisomerase IIβ, bacterial gyrase, and topoisomerase IV.
- Type IIB topoisomerases, which include topoisomerase VI found in archaea.
| Topoisomerase[18] | Subfamily type | Function | Multimericity | Metal dependence | ATP dependence | Single- or double-stranded cleavage? | Cleavage polarity | Change in link number (L) |
|---|---|---|---|---|---|---|---|---|
| Topoisomerase I
(E. coli) |
Type IA | Removes (-), but not (+) supercoils | Monomer | Yes (Mg2+) | No | SS | 5' | ±1 |
| Topoisomerase III
(E. coli) |
Removes (-), but not (+) supercoils; overlapping function with topoisomerase IV | |||||||
| Topoisomerase IIIα
(H. sapiens) |
Removes (-), but not (+) supercoils; assists in the unlinking of precatenanes in cellular DNA replication; can catalyze the knotting, unknotting, and interlinking of single-stranded circles as well as the knotting, unknotting, catenation, and decatenation of gapped or nicked duplex DNA circles. | |||||||
| Topoisomerase IIIβ
(H. sapiens) |
Unknown function | |||||||
| Reverse DNA gyrase
(E. coli) |
Removes (-), but not (+) supercoils | Heterodimer | ||||||
| Reverse DNA gyrase
(Archaea) |
Removes (-), but not (+) supercoils | |||||||
| Topoisomerase I
(H. sapiens) |
Type IB | Remove (+) and (-) supercoils; relaxes compensatory (-) supercoils; generates right-handed solenoidal supercoils; supports fork movement during replication; thought to be similar in structure to tyrosine recombinases. | Monomer | No | No | SS | 3' | ±1 |
| Topoisomerase V
(Archaea)[28] |
Type IC | Relaxes (+) and (-) supercoils. Involved in DNA repair. In UniProt: F1SVL0, Q977W1. | Monomer | No | No | SS | 3' | ±1 |
| Topoisomerase II / DNA gyrase
(E. coli) |
Type IIA | Generates (-) supercoils (the only topoisomerase known to do this) | Heterotetramer | Yes (Mg2+) | Yes | DS | 5' | ±2 |
| Topoisomerase IV
(E. coli) |
Relaxes (-) supercoils; role in decatenation | Heterotetramer | ||||||
| Topoisomerase IIα
(H. sapiens) |
Essential; Unlinks intertwined daughter duplexes in replication; contributes to DNA relaxation during transcription | Homodimer | ||||||
| Topoisomerase IIβ
(H. sapiens) |
Role in suppressing recombination or supporting transcription in neurons | Homodimer | ||||||
| Topoisomerase VI
(Archaea) |
Type IIB | Relaxes (+) and (-) supercoils; responsible for decatenating replication intermediates; May be exclusive to the archaea. | Heterotetramer | Yes (Mg2+) | Yes | DS | 5' | ±2 |
Both type I and type II topoisomerases change the linking number (L) of DNA. Type IA topoisomerases change the linking number by one, type IB and type IC topoisomerases change the linking number by any integer, whereas type IIA and type IIB topoisomerases change the linking number by two.[citation needed]
DNA topoisomerase type I family
DNA topoisomerase type I family consists of two subfamilies; type IA and type IB. Type IA DNA topoisomerase amongst various organisms generally share the following properties: All of the enzymes are monomers. The enzyme shares a covalent interaction of a 5' phosphodiester bond at its tyrosine active site with the end of a DNA strand. The mechanism of relaxation of supercoiling requires magnesium(II). In plasmid DNA, the negative supercoils produced can be substrates for the relaxation mechanism, a process which does not go to completion. Type IA also requires an exposed single-stranded region within the DNA substrate. The linking number of DNA changes with relaxation. Type IA topoisomerase can catalyze catenation, decatenation, knotting and unknotting of the DNA.[16]
There are three classes within the subfamily of type IB topoisomerase: topoisomerase I in eukaryotes, topoisomerase V in prokaryotes, and the poxvirus topoisomerase. Type IB subfamily topoisomerases are generally classified by their ability to relax both negative and positive supercoils, and the relaxation mechanism goes to completion (unlike type IA). They form a covalent interaction through the tyrosine active site on the enzyme and the 3’ phosphate on the DNA strand. The relaxation mechanism does not require magnesium(II). Both type IA and type IB topoisomerases, within the Type I family, have very distinct differences in their properties.
DNA topoisomerase type II family
Type II family of topoisomerases share general features and properties that make them distinguishable from the type I family. All type II DNA topoisomerases are dimers. They bind to a duplex DNA and cleave both strands, staggering four bases. Cleavage is done by a covalent interaction between each dimer subunit to the 5’ phosphate on the DNA, creating a phosphotyrosine bond. The reaction pulls the two ends of the cleaved DNA apart – this is called the gated (G-) segment. The transported (T-) segment, a region on the same or a different DNA duplex, is passed through G-segment. This changes the linking number when the DNA is circular (plasmid). The relaxation mechanism requires magnesium(II) and hydrolysis of ATP. The active site containing amino acid tyrosines have a helix-turn-helix motif, which collaborates with acidic residues to activate catalysis. Prokaryotic type II topoisomerases are heterotetrameric. Eukaryotic type II topoisomerases are homodimeric. Type II are also better at DNA relaxation, instead of decatenation, meaning they are better enzymes for relieving topological stresses in linear DNA versus circular DNA.
See also
- DNA topology
- Supercoil
- Type I topoisomerase
- Type II topoisomerase
- Topoisomerase I
- Topoisomerase IIα
- Topoisomerase IIβ
- Topoisomerase IIIα
- Topoisomerase IIIβ
References
- ^ McKie, Shannon J.; Neuman, Keir C.; Maxwell, Anthony (2021-04). "DNA topoisomerases: Advances in understanding of cellular roles and multi‐protein complexes via structure‐function analysis". BioEssays. 43 (4): 2000286. doi:10.1002/bies.202000286. ISSN 0265-9247.
{{cite journal}}: Check date values in:|date=(help) - ^ Sutormin, Dmitry A.; Galivondzhyan, Alina Kh.; Polkhovskiy, Alexander V.; Kamalyan, Sofia O.; Severinov, Konstantin V.; Dubiley, Svetlana A. (2021-03-15). "Diversity and Functions of Type II Topoisomerases". Acta Naturae. 13 (1): 59–75. doi:10.32607/actanaturae.11058. ISSN 2075-8251. PMC 8084294. PMID 33959387.
- ^ Wang, James C. (1971-02). "Interaction between DNA and an Escherichia coli protein ω". Journal of Molecular Biology. 55 (3): 523–IN16. doi:10.1016/0022-2836(71)90334-2. ISSN 0022-2836.
{{cite journal}}: Check date values in:|date=(help) - ^ Champoux, James J.; Dulbecco, Renato (1972-01). "An Activity from Mammalian Cells That Untwists Superhelical DNA—A Possible Swivel For DNA Replication". Proceedings of the National Academy of Sciences. 69 (1): 143–146. doi:10.1073/pnas.69.1.143. ISSN 0027-8424.
{{cite journal}}: Check date values in:|date=(help) - ^ Gellert, M; Mizuuchi, K; O'Dea, M H; Nash, H A (1976-11). "DNA gyrase: an enzyme that introduces superhelical turns into DNA". Proceedings of the National Academy of Sciences. 73 (11): 3872–3876. doi:10.1073/pnas.73.11.3872. ISSN 0027-8424.
{{cite journal}}: Check date values in:|date=(help) - ^ Sugino, Akio; Peebles, Craig L.; Kreuzer, Kenneth N.; Cozzarelli, Nicholas R. (1977-11). "Mechanism of action of nalidixic acid: Purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme". Proceedings of the National Academy of Sciences. 74 (11): 4767–4771. doi:10.1073/pnas.74.11.4767. ISSN 0027-8424.
{{cite journal}}: Check date values in:|date=(help); line feed character in|title=at position 55 (help) - ^ Baldi, M (1980-06). "In vitro catenation and decatenation of DNA and a novel eucaryotic ATP-dependent topoisomerase". Cell. 20 (2): 461–467. doi:10.1016/0092-8674(80)90632-7. ISSN 0092-8674.
{{cite journal}}: Check date values in:|date=(help) - ^ Liu, Leroy F.; Liu, Chung-Cheng; Alberts, Bruce M. (1979-10). "T4 DNA topoisomerase: a new ATP-dependent enzyme essential for initiation of T4 bacteriophage DNA replication". Nature. 281 (5731): 456–461. doi:10.1038/281456a0. ISSN 0028-0836.
{{cite journal}}: Check date values in:|date=(help) - ^ Stetler, G L; King, G J; Huang, W M (1979-08). "T4 DNA-delay proteins, required for specific DNA replication, form a complex that has ATP-dependent DNA topoisomerase activity". Proceedings of the National Academy of Sciences. 76 (8): 3737–3741. doi:10.1073/pnas.76.8.3737. ISSN 0027-8424.
{{cite journal}}: Check date values in:|date=(help) - ^ Bates, Andrew D. (2005). DNA topology. Anthony Maxwell (2nd ed ed.). Oxford: Oxford University Press. ISBN 978-0-19-154658-7. OCLC 64239232.
{{cite book}}:|edition=has extra text (help) - ^ WATSON, J. D.; CRICK, F. H. C. (1953). "Genetical Implications of the Structure of Deoxyribonucleic Acid". Nature. 171 (4361): 964–967. doi:10.1038/171964b0. ISSN 0028-0836.
- ^ WATSON, J. D.; CRICK, F. H. C. (1953-04-25). "Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid". Nature. 171 (4356): 737–738. doi:10.1038/171737a0. ISSN 0028-0836.
- ^ FRANKLIN, ROSALIND E.; GOSLING, R. G. (1953). "Evidence for 2-Chain Helix in Crystalline Structure of Sodium Deoxyribonucleate". Nature. 172 (4369): 156–157. doi:10.1038/172156a0. ISSN 0028-0836.
- ^ CAIRNS, JOHN (1989), "The Bacterial Chromosome and its Manner of Replication as seen by Autoradiography", Molecular Biology, Elsevier, pp. 177–185, retrieved 2022-04-09
- ^ Delbrück, M. (1954). "ON THE REPLICATION OF DESOXYRIBONUCLEIC ACID (DNA)". Proceedings of the National Academy of Sciences. 40 (9): 783–788. doi:10.1073/pnas.40.9.783. ISSN 0027-8424.
- ^ a b "DNA TOPOISOMERASES: Structure, Function, and Mechanism". Annual Review of Biochemistry. 70: 369–413. 2001. doi:10.1146/annurev.biochem.70.1.369. PMID 11395412.
{{cite journal}}: Unknown parameter|authors=ignored (help) - ^ Hogan CM (2010). "Deoxyribonucleic acid". In Draggan S, Cleveland C (eds.). Encyclopedia of Earth. Washington DC: National Council for Science and the Environment.
- ^ a b Champoux JJ (2001). "DNA topoisomerases: structure, function, and mechanism". Annual Review of Biochemistry. 70: 369–413. doi:10.1146/annurev.biochem.70.1.369. PMID 11395412.
- ^ Vologodskii A (September 2011). "Unlinking of supercoiled DNA catenanes by type IIA topoisomerases". Biophysical Journal. 101 (6): 1403–11. Bibcode:2011BpJ...101.1403V. doi:10.1016/j.bpj.2011.08.011. PMC 3177067. PMID 21943421.
- ^ a b Sobek S, Boege F (August 2014). "DNA topoisomerases in mtDNA maintenance and ageing". Experimental Gerontology. 56: 135–41. doi:10.1016/j.exger.2014.01.009. PMID 24440386.
- ^ Tsai HZ, Lin RK, Hsieh TS (April 2016). "Drosophila mitochondrial topoisomerase III alpha affects the aging process via maintenance of mitochondrial function and genome integrity". Journal of Biomedical Science. 23: 38. doi:10.1186/s12929-016-0255-2. PMC 4828762. PMID 27067525.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ McCarthy D, Minner C, Bernstein H, Bernstein C (October 1976). "DNA elongation rates and growing point distributions of wild-type phage T4 and a DNA-delay amber mutant". Journal of Molecular Biology. 106 (4): 963–81. doi:10.1016/0022-2836(76)90346-6. PMID 789903.
- ^ Mufti S, Bernstein H (October 1974). "The DNA-delay mutants of bacteriophage T4". Journal of Virology. 14 (4): 860–71. doi:10.1128/JVI.14.4.860-871.1974. PMC 355592. PMID 4609406.
- ^ Miskimins R, Schneider S, Johns V, Bernstein H (June 1982). "Topoisomerase involvement in multiplicity reactivation of phage T4". Genetics. 101 (2): 157–77. doi:10.1093/genetics/101.2.157 (inactive 28 February 2022). PMC 1201854. PMID 6293912.
{{cite journal}}: CS1 maint: DOI inactive as of February 2022 (link) - ^ "The dynamic interplay between DNA topoisomerases and DNA topology". Biophysical Reviews. 8 (Suppl 1): 101–111. November 2016. doi:10.1007/s12551-016-0240-8. PMC 5418509. PMID 28510219.
{{cite journal}}: Unknown parameter|authors=ignored (help) - ^ Pommier Y, Leo E, Zhang H, Marchand C (May 2010). "DNA topoisomerases and their poisoning by anticancer and antibacterial drugs". Chemistry & Biology. 17 (5): 421–33. doi:10.1016/j.chembiol.2010.04.012. PMC 7316379. PMID 20534341.
- ^ Wang JC (April 1991). "DNA topoisomerases: why so many?". The Journal of Biological Chemistry. 266 (11): 6659–62. doi:10.1016/S0021-9258(20)89545-3. PMID 1849888.
- ^ a b Baker NM, Rajan R, Mondragón A (February 2009). "Structural studies of type I topoisomerases". Nucleic Acids Research. 37 (3): 693–701. doi:10.1093/nar/gkn1009. PMC 2647283. PMID 19106140.
Further reading
- Wang JC (2009). Untangling the Double Helix: DNA entanglement and the action of the DNA topoisomerases. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. p. 245. ISBN 978-0-87969-879-9.
External links
- DNA+Topoisomerases at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
