EngA RNA motif
Appearance
| engA | |
|---|---|
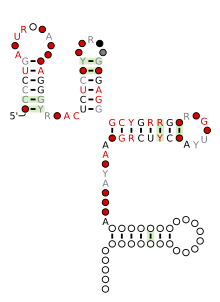 Consensus secondary structure and sequence conservation of engA RNA | |
| Identifiers | |
| Symbol | engA |
| Rfam | RF02972 |
| Other data | |
| RNA type | Cis-reg |
| SO | SO:0005836 |
| PDB structures | PDBe |
The engA RNA motif is a conserved RNA structure that was discovered by bioinformatics.[1] engA motifs are found in bacteria within the genus Prevotella.
engA motif RNAs likely function as cis-regulatory elements, in view of their positions upstream of protein-coding genes. They are consistently located upstream of genes encoding GTPases, many of which are annotated as encoding the protein EngA. EngA participates in ribosome stability and assembly.
References
[edit]- ^ Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR (October 2017). "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. doi:10.1093/nar/gkx699. PMC 5737381. PMID 28977401.
