Cytochrome c oxidase

The enzyme cytochrome c oxidase or Complex IV (PDB: 2OCC, EC 1.9.3.1) is a large transmembrane protein complex found in bacteria and the mitochondrion.
Function
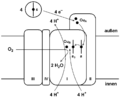
It is the last protein in the electron transport chain. It receives an electron from each of four cytochrome c molecules, and transfers them to one oxygen molecule, converting molecular oxygen to two molecules of water. In the process, it translocates four protons, helping to establish a chemiosmotic potential that the ATP synthase then uses to synthesize ATP.
Reaction
Summary reaction:
- 4 Fe2+-cytochrome c + 8 H+in + O2 → 4 Fe3+-cytochrome c + 2 H2O + 4 H+out
Structure
The complex is a large lipoprotein composed of several metal prosthetic sites and 13 protein subunits in mammals. In mammals, ten subunits are nuclear in origin and three are synthesized mitochondrially. The complex contains two hemes, the a and a3 hemes, and two copper centers, the CuA and CuB centers. In fact, the heme a3 and CuB are a binuclear center that is the site of oxygen reduction. The mechanism of action of this large complex is still an active research topic.
Crystallographic studies of cytochrome c oxidase show an unusual post translational modification, linking C6 of Tyr(244) and the ε-N of His(240) (bovine enzyme numbering). This cross-linked tyrosine has been proposed to serve as a hydrogen donor, providing a proton and an electron to cleave the dioxygen bond. Other redox reactive amino acids used for catalysis include glycine, cysteine, tyrosine, tryptophan and a similarly modified tyrosine (cross-linked to cysteine).
Inhibition
Cyanide, sulfide, azide and carbon monoxide[1] all bind to Cytochrome c Oxidase, thus inhibiting the protein from functioning which results in chemical suffocation of cells.
Genetic Defects and Disorders
Defects involving genetic mutations altering cytochrome c oxidase (COX) functionality or structure can rsult in severe, often fatal metabolic disorders. Such disorders usually manifest in early childhood and predominantly affect tissues with high energetic demands (brain, heart, muscle). Among the many classified mitochondrial diseases, those involving dysfunctional COX assembly are thought to be the most severe[2]
The vast majority of COX disorders are linked to mutations in nuclearly encoded proteins referred to as assembly factors, or assembly proteins. These assembly factors contribute to COX structure and functionality and are involved in sevral essential processess including transcription and translation of mitochondrial encoded subunits, processing of preproteins and membrane insertion, and cofactor biosynthesis and incorporation. [3]
Currently, mutations have been identified in six COX assembly factors: SURF1, SCO1, SCO2, COX10, COX15, and LRPPRC.[4] Mutations in these proteins can result in altered functionality of sub-complex assembly, copper transport, or translational regulation. Each gene mutation is associated with the etiology of a specific disease, with some having implications in multiple disorders. Disorders involving dysfunctional COX assembly via gene mutations include Leigh syndrome, cardiomyopathy, leukodystrophy, anemia, and sensorineural deafness.[5][6]
Additional images
-
ETC
-
Complex IV
References
- ^ Alonso J, Cardellach F, López S, Casademont J, Miró O (2003). "Carbon monoxide specifically inhibits cytochrome c oxidase of human mitochondrial respiratory chain". Pharmacol Toxicol. 93 (3): 142–6. PMID 12969439.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pecina, P., Houstkova, H., Hansikova, H., Zeman, J., Houstek, J. (2004). Genetic Defects of Cytocrhome c Oxidase Assembly. Physiol. Res. 53(Suppl. 1): S213-S223.
- ^ Zee, J.M., and Glerum, D.M. (2006). Defects in cytochrome oxidase assembly in humans: lessons from yeast. Biochem. Cell Biol. 84: 859-869.
- ^ Zee, J.M., and Glerum, D.M. (2006). Defects in cytochrome oxidase assembly in humans: lessons from yeast. Biochem. Cell Biol. 84: 859-869.
- ^ Pecina, P., Houstkova, H., Hansikova, H., Zeman, J., Houstek, J. (2004). Genetic Defects of Cytocrhome c Oxidase Assembly. Physiol. Res. 53(Suppl. 1): S213-S223.
- ^ Zee, J.M., and Glerum, D.M. (2006). Defects in cytochrome oxidase assembly in humans: lessons from yeast. Biochem. Cell Biol. 84: 859-869.
External links
- The Cytochrome Oxidase home page at Rice University
- Interactive Molecular model of cytochrome c oxidase (Requires MDL Chime)
- UMich Orientation of Proteins in Membranes families/superfamily-4
- Cytochrome-c+Oxidase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)