User:Cellular Biochemistry II/SopE
| SopE | |||||||
|---|---|---|---|---|---|---|---|
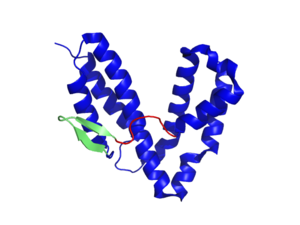 Crystal structure of the catalytic domain of SopE | |||||||
| Identifiers | |||||||
| Symbol | SopE | ||||||
| Alt. symbols | STY4609 | ||||||
| NCBI gene | 1250812 | ||||||
| PDB | 1GZS | ||||||
| UniProt | O52623 | ||||||
| |||||||
SopE, Salmonella outer protein E, is a protein encoded by the genome of Salmonella, a genus of gram-negative motile enterobacteria that can be found in animals, humans and non-living habitats. During the entry of Salmonella typhimurium into mammalian intestinal epithelial cells, the bacterial SopE is, among other proteins, injected via a conserved type III secretion system (T3SS) into the cytosol of the host cells. SopE is a Rho guanine nucleotide exchange factor (GEF) for Cdc42 and Rac and activates these GTPases in the host cell to induce membrane ruffling and actin cytoskeleton rearrangement supporting the internalization of Salmonella into to host cell. [1] [2] [3]
Structure
[edit]

SopE is a monomeric protein with 240 amino acids and a molecular weight of 26.6 kDa. This GEF for Cdc42 and Rac is unusual, as the catalytic domain (residues 78-240) consists of six α-helices in two three-helix bundles and a two stranded β-sheet followed by an 8 amino acid long peptide segment. This peptide contains the catalytic core of SopE, a 166GAGA169 motif (cf Figure 2) [4]. Thus SopE does not exhibit the for mammalian Rho-GEFs typical protein signatures e.g. the catalytical DH (Dbl homology) or docker domains; instead only 4 amino acids form its catalytical core site. The structural analysis of the SopE-Cdc42 complex revealed: By inserting the 166GAGA169-loop between the in nucleotide-binding involved switch I and II regions of Cdc42, it induces major conformational changes thereby reducing the affinity for GDP. These changes include especially the extrusion of Phe28Cdc42, which normally contributes to nucleotide-binding in switch I, as well as a peptide flip of Ala59Cdc42 in switch II, which leads to the blockage of the Mg2+ binding site. This might be an important step in guanine nucleotide release since Mg2+ is important for high-affinity binding of guanine nucleotides to GTPases. Interestingly, similar conformational changes in these switch regions have been observed in other eukaryotic Rho family proteins leading to the assumption that SopE mimics the action of Dbl homology-Pleckstrin homology (DH-PH) domains found in Rho-specific eukaryotic GEFs. [5] [6]
Regulation of SopE expression
[edit]The bacterial T3SS consists of structural, secreted and regulatory proteins, so called chaperons. There are bacterial proteins that regulate the stability of the "to-be-secreted" proteins. The transcription activator InvF, a member of the AraC/XylS family [7] [8] binds with its C-terminal helix-turn-helix (HTH) motif to e.g. the sopE promoter. In addition a regulatory protein, a so called type III secretion chaperone, named SicA, also activates the transcription of SopE [9] [10]. Some Experiments verify the thesis that SicA interacts with InvF, but the function is not resolved. [11].
Function
[edit]SopE is absent from most Salmonella typhimurium strains. But the 69% similar protein SopE2 is expressed in all S. typhimurium strains and seems to act similarly to SopE.
SopE mediated cytoskeletal and nuclear responses are dependent on Cdc42 and Rac-1
[edit]SopE plays a crucial role in the Salmonella trigger mechanism, which encompasses the way a bacterium enters the host cell. SopE is injected into the host cell's cytoplasm and induces rearrangement of the host actin cytoskeleton due to direct activation of both Cdc42 and Rac1, host proteins. [12]
The contact between the bacteria and the host cell is mediated by the T3SS, which allows direct activation of cytoskeletal components by delivered bacterial effectors.
[13]
This contact process can be divided into 4 steps, where SopE is a main actor at the third step.
- Pre-interaction stage: effector molecules are stored in the bacterial cytoplasm in association with dedicated chaperones to avoid their degradation
- Interaction stage: formation of a signaling platform, where a translocon pore is formed.
- Formation of a macropinocytic pocket. The cell surface near the translocon pore is massively rearranged by formation of filopodial and lamelliopodial structures. The translocated SopE protein, which has a GEF activity, acts as exchange factor for Cdc42 and Rac1. Thus actin polymerization is massively boosted. SopE is then rapidly degraded through a proteasome dependent pathway.
- In the last step the newly polymerized actin gets depolymerisation and the macropinocytic pocket closed. [14]
SopE induces also the activation of the MAP Kinase JNK, in a Cdc24 and Rac-1 dependent manner, leading to cytoskeletal and nuclear responses.
InvB is a chaperone of SopE
[edit]The chaperone InvB is required for secretion and translocation of SopE. SopE directly interacts with InvB through a domain located at its amino terminus. The chaperone and its substrate are maintained in two separate genetic elements, a pathogenicity island (SPI-1) and an integrated bacteriophage. Both elements were presumably independent from each other horizontally acquired throughout evolution. Therefore, InvB serves as a chaperone for two or perhaps even three secreted proteins that are genetically unlinked. [15]
SopE stimulates GDP/GTP Nucleotide Exchange in Members of the Rho Subfamily of GTPase in vitro
[edit]SopE-induced cellular responses depend on small Rho-like GTPases. SopE can activate several members of the Rho subfamily of GTPases in vitro and has been shown to induce both binding of the GTP-analogue [35S]GTP-γ-S to GDP-loaded Rac-1 as well as the release of [3H]GDP from Rac-1. [16]
Antagonist of SopE effects
[edit]After bacterial internalization, the cell regains its normal architecture. SptP, also a secreted protein of the T3SS acts as a GTPase-activating protein (GAP) for Cdc42 and Rac-1 and is responsible for the reversal of the actin cytoskeleton rearrangement in the host cell [17].
Efficiency
[edit]Cdc42G12V in complex with fluorescent GDP was used for biophysical analysis to determine the kinetic data of SopE78-240 from Salmonella thyphimurium; kcat = 0.95 ± 0.006 s-1, Km = 4.5 ± 0.9 μM and kcat /Km = 2.1 × 105 M-1s-1. SopE has a high catalytic efficiency, which is usual for a protein that induces a large conformational change [18].
References
[edit]- ^
Ehrbar K, Mirold S, Friebel A, Stender S, Hardt WD (2002). "Characterization of effector proteins translocated via the SPI1 type III secretion system of Salmonella typhimurium". Int J Med Microbiol. 291(6-7): 479–485.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Zhou D, Galán J (2001). "Salmonella entry into host cells: the work in concert of type III secreted effector proteins". Microbes and Infection. 3(14-15): 1293–1298.
- ^
Aktories K, Schmidt G, Just I (2000). "Rho GTPases as targets of bacterial protein toxins". Biol Chem. 381(5-6): 421–426.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^
Buchwald G., Friebel A., Galan J.E., Hardt W.-D., Wittinghofer A., Scheffzek K. (2002). "Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE". EMBO Journal. 21: 3286–3295.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^
Schlumberger M.C., Friebel A., Buchwald G., Scheffzek K., Wittinghofer A., Hardt W.-D. (2003). "Amino acids of the bacterial toxin SopE involved in G nucleotide exchange on Cdc42". J Biol Chem. 278: 27149–27159.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Erickson JW and Cerione RA (2004). "Structural Elements, Mechanism, and Evolutionary Convergence of Rho Protein−Guanine Nucleotide Exchange Factor Complexes". Biochemistry. 43(4): 837–842.
- ^ Eichelberg K., Galan J.E (1999). "Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA". Infection and Immunity. 67(8): 4099–4105.
- ^
Mills D.M., Bajaj V., Lee C.A. (1995). "A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome". Mol Microbiol. 15(4): 749–759.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Darwin K.H., Miller V.L. (2000). "Putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes". Mol Microbiol. 35: 949–959.
- ^ Tucker S.C., Galan J.E. (2000). "Complex function for SicA, a Salmonella enterica serovar typhimurium type III secretion associated chaperone". Journal of Bacteriology. 182: 2262–2268.
- ^ Darwin K.H., Miller V.L. (2001). "Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium". EMBO J. 20(8): 1850–1862.
- ^
Friebel A., Ilchmann H., Aepfelbacher M., Ehrbar K., Machleidt W., Hardt W.-D. (2001). "SopE and SopE2 from Salmonella typhimurium activate different sets of RhoGTPases of the host cell". J Biol Chem. 276(36): 34035–34040.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ P. J. Sansonetti (2001). "Rupture, invasion and inflammatory destruction of the intestinal barrier by Shigella, making sense of prokaryote-eukaryote cross-talks". FEMS Microbiol Rev. 25(1): 3–14.
- ^ J. E. Galan (2001). "Salmonella Interactions with host cells: Type III Secretions system at work". Annu Rev Cell Dev Biol. 17: 53–86.
- ^ Lee S.H., Galan J.E. (2003). "InvB is a type III secretion-associated chaperone for the Salmonella enterica effector protein SopE". Journal of Bacteriology. 185(24): 7279–7284.
- ^
W D Hardt, L M Chen, K E Schuebel, X R Bustelo, J E Galán. "S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells". Cell. 93(5): 815–26.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Fu Y., Galan J.E. (1999). "A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion". Nature. 401.
- ^
Rudolph M.G., Weise C., Mirold S., Hillenbrand B., Bader B., Wittinghofer A., Hardt W.-D. (1999). "Biochemical Analysis of SopE from Salmonella typhimurium, a Highly Efficient Guanosine Nucleotide Exchange Factor for RhoGTPases". J Biol Chem. 274(43): 30501–30509.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
