DNA polymerase
| DNA-directed DNA polymerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 3D structure of the DNA-binding helix-turn-helix motifs in human DNA polymerase beta (based on pdb file 7ICG) | |||||||||
| Identifiers | |||||||||
| EC no. | 2.7.7.7 | ||||||||
| CAS no. | 9012-90-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
The DNA polymerases are enzymes that create DNA molecules by assembling nucleotides, the building blocks of DNA. These enzymes are essential to DNA replication and usually work in pairs to create two identical DNA strands from a single original DNA molecule. During this process, DNA polymerase “reads” the existing DNA strands to create two new strands that match the existing ones.
Every time a cell divides, DNA polymerase is required to help duplicate the cell’s DNA, so that a copy of the original DNA molecule can be passed to each of the daughter cells. In this way, genetic information is transmitted from generation to generation.
Before replication can take place, an enzyme called helicase unwinds the DNA molecule from its tightly woven form. This opens up or “unzips” the double-stranded DNA to give two single strands of DNA that can be used as templates for replication.
History
In 1956, Arthur Kornberg and colleagues discovered the enzyme DNA polymerase I, also known as Pol I, in Escherichia coli. They described the DNA replication process by which DNA polymerase copies the base sequence of a template DNA strand. Subsequently, in 1959, Kornberg was awarded the Nobel Prize in Physiology or Medicine for this work.[1] DNA polymerase II was also discovered by Kornberg and Malcolm E. Gefter in 1970 while further elucidating the role of Pol I in E. coli DNA replication.[2]
Function

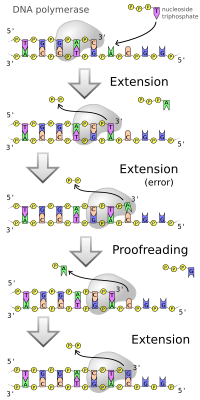
The main function of DNA polymerase is to make DNA from nucleotides, the building blocks of DNA. The DNA copies are created by the pairing of nucleotides to bases present on each strand of the original DNA molecule. This pairing always occurs in specific combinations, with cytosine and guanine and thymine and adenine forming two separate pairs respectively.
When creating DNA, DNA polymerase can add free nucleotides only to the 3' end of the newly forming strand. This results in elongation of the newly forming strand in a 5'-3' direction. No known DNA polymerase is able to begin a new chain (de novo); it can only add a nucleotide onto a pre-existing 3'-OH group, and therefore needs a primer at which it can add the first nucleotide. Primers consist of RNA or DNA bases (or both). In DNA replication, the first two bases are always RNA, and are synthesized by another enzyme called primase. An enzyme known as a helicase is required to unwind DNA from a double-strand structure to a single-strand structure to facilitate replication of each strand consistent with the semiconservative model of DNA replication.
It is important to note that the directionality of the newly forming strand (the daughter strand) is opposite to the direction in which DNA polymerase moves along the template strand. Since DNA polymerase requires a free 3' OH group for initiation of synthesis, it can synthesize in only one direction by extending the 3' end of the preexisting nucleotide chain. Hence, DNA polymerase moves along the template strand in a 3'-5' direction, and the daughter strand is formed in a 5'-3' direction. This difference enables the resultant double-strand DNA formed to be composed of two DNA strands that are antiparallel to each other.
The function of DNA polymerase is not quite perfect, with the enzyme making about one mistake for every billion base pairs copied. Error correction is a property of some, but not all, DNA polymerases. This process corrects mistakes in newly synthesized DNA. When an incorrect base pair is recognized, DNA polymerase moves backwards by one base pair of DNA. The 3'-5' exonuclease activity of the enzyme allows the incorrect base pair to be excised (this activity is known as proofreading). Following base excision, the polymerase can re-insert the correct base and replication can continue forwards. This preserves the integrity of the original DNA strand that is passed onto the daughter cells.
Structure
The known DNA polymerases have highly conserved structure, which means that their overall catalytic subunits vary very little from species to species, independent of their domain structures. The shape can be described as resembling a right hand with thumb, finger, and palm domains. The palm domain appears to function in catalyzing the transfer of phosphoryl groups in the phosphoryl transfer reaction. DNA is palm when the enzyme is active. This reaction is believed to be catalyzed by a two metal ion mechanism. The finger domain functions to bind the nucleotide triphosphate with the template base. The thumb domain plays a potential role in the processivity, translocation, and positioning of the DNA.[3] Conserved structures usually indicate important, irreplaceable functions of the cell, the maintenance of which provides evolutionary advantages.
Processivity
DNA polymerase’s rapid catalysis is due to its processive nature. Processivity is a characteristic of enzymes that function on polymeric substrates. In the case of DNA polymerase, the degree of processivity refers to the average number of nucleotides added each time the enzyme binds a template. The average DNA polymerase requires about one second locating and binding a primer/template junction. Once it is bound, a nonprocessive DNA polymerase adds nucleotides at a rate of one nucleotide per second.[4]: 207–208 Processive DNA polymerases, however, add multiple nucleotides per second, drastically increasing the rate of DNA synthesis. The degree of processivity is directly proportional to the rate of DNA synthesis. The rate of DNA synthesis in a living cell was first determined as the rate of phage T4 DNA elongation in phage infected E. coli. During the period of exponential DNA increase at 37 °C, the rate was 749 nucleotides per second.[5]
DNA polymerase’s ability to slide along the DNA template allows increased processivity. There is a dramatic increase in processivity at the replication fork. This increase is facilitated by the DNA polymerase’s association with proteins known as the sliding DNA clamp. The clamps are multiple protein subunits associated in the shape of a ring. Using the hydrolysis of ATP, a class of proteins known as the sliding clamp loading proteins open up the ring structure of the sliding DNA clamps allowing binding to and release from the DNA strand. Protein-protein interaction with the clamp prevents DNA polymerase from diffusing from the DNA template, thereby ensuring that the enzyme binds the same primer/template junction and continues replication.[4]: 207–208 DNA polymerase changes conformation, increasing affinity to the clamp when associated with it and decreasing affinity when it completes the replication of a stretch of DNA to allow release from the clamp.
Variation across species
| DNA polymerase family A | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 c:o6-methyl-guanine pair in the polymerase-2 basepair position | |||||||||
| Identifiers | |||||||||
| Symbol | DNA_pol_A | ||||||||
| Pfam | PF00476 | ||||||||
| InterPro | IPR001098 | ||||||||
| SMART | - | ||||||||
| PROSITE | PDOC00412 | ||||||||
| SCOP2 | 1dpi / SCOPe / SUPFAM | ||||||||
| |||||||||
| DNA polymerase family B | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of rb69 gp43 in complex with dna containing thymine glycol | |||||||||
| Identifiers | |||||||||
| Symbol | DNA_pol_B | ||||||||
| Pfam | PF00136 | ||||||||
| Pfam clan | CL0194 | ||||||||
| InterPro | IPR006134 | ||||||||
| PROSITE | PDOC00107 | ||||||||
| SCOP2 | 1noy / SCOPe / SUPFAM | ||||||||
| |||||||||
| DNA polymerase type B, organellar and viral | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 phi29 dna polymerase, orthorhombic crystal form, ssdna complex | |||||||||
| Identifiers | |||||||||
| Symbol | DNA_pol_B_2 | ||||||||
| Pfam | PF03175 | ||||||||
| Pfam clan | CL0194 | ||||||||
| InterPro | IPR004868 | ||||||||
| |||||||||
Based on sequence homology, DNA polymerases can be further subdivided into seven different families: A, B, C, D, X, Y, and RT.
Some viruses also encode special DNA polymerases, such as Hepatitis B virus DNA polymerase. These may selectively replicate viral DNA through a variety of mechanisms. Retroviruses encode an unusual DNA polymerase called reverse transcriptase, which is an RNA-dependent DNA polymerase (RdDp). It polymerizes DNA from a template of RNA.
| Family | Types of DNA polymerase | Species | Examples |
|---|---|---|---|
| A | Replicative and Repair Polymerases | Eukaryotic and Prokaryotic | T7 DNA polymerase, Pol I, and DNA Polymerase γ |
| B | Replicative and Repair Polymerases | Eukaryotic and Prokaryotic | Pol II, Pol B, Pol ζ, Pol α, δ, and ε |
| C | Replicative Polymerases | Prokaryotic | Pol III |
| D | Replicative Polymerases | Euryarchaeota | Not well-characterized |
| X | Replicative and Repair Polymerases | Eukaryotic | Pol β, Pol σ, Pol λ, Pol μ, and Terminal deoxynucleotidyl transferase |
| Y | Replicative and Repair Polymerases | Eukaryotic and Prokaryotic | Pol ι (iota), Pol κ (kappa), Pol IV, and Pol V |
| RT | Replicative and Repair Polymerases | Viruses, Retroviruses, and Eukaryotic | Telomerase, Hepatitis B virus |
Prokaryotic DNA polymerase
Pol I
Prokaryotic Family A polymerases include the DNA polymerase I (Pol I) enzyme, which is encoded by the polA gene and ubiquitous among prokaryotes. This repair polymerase is involved in excision repair with 3'-5' and 5'-3' exonuclease activity and processing of Okazaki fragments generated during lagging strand synthesis.[6] Pol I is the most abundant polymerase accounting for >95% of polymerase activity in E. coli, yet cells lacking Pol I have been found suggesting Pol I activity can be replaced by the other four polymerases. Pol I adds ~15-20 nucleotides per second, thus showing poor processivity. Instead, Pol I starts adding nucleotides at the RNA primer:template junction known as the origin of replication (ori). Approximately 400 bp downstream from the origin, the Pol III holoenzyme is assembled and takes over replication at a highly processive speed and nature.[7]
Pol II
DNA polymerase II, a Family B polymerase, is a polB gene product also known as DinA. Pol II has 3'-5' exonuclease activity and participates in DNA repair, replication restart to bypass lesions, and its cell presence can jump from ~30-50 copies per cell to ~200-300 during SOS induction. Pol II is also thought to be a backup to Pol III as it can interact with holoenzyme proteins and assume a high level of processivity. The main role of Pol II is thought to be the ability to direct polymerase activity at the replication fork and helped stalled Pol III bypass terminal mismatches.[8]
Pol III
DNA polymerase III holoenzyme is the primary enzyme involved in DNA replication in E. coli and belongs to Family C polymerases. It consists of three assemblies: the pol III core, the beta sliding clamp processivity factor and the clamp-loading complex. The core consists of three subunits - α, the polymerase activity hub, ɛ, exonucleolytic proofreader, and θ, which may act as a stabilizer for ɛ. The holoenzyme contains two cores, one for each strand, the lagging and leading.[9] The beta sliding clamp processivity factor is also present in duplicate, one for each core, to create a clamp that encloses DNA allowing for high processivity.[10] The third assembly is a seven-subunit (τ2γδδ′χψ) clamp loader complex. Recent research has classified Family C polymerases as a subcategory of Family X with no eukaryotic equivalents.[11]
Pol IV
In E. coli, DNA polymerase IV (Pol 4) is an error-prone DNA polymerase involved in non-targeted mutagenesis.[12] Pol IV is a Family Y polymerase expressed by the dinB gene that is switched on via SOS induction caused by stalled polymerases at the replication fork. During SOS induction, Pol IV production is increased tenfold and one of the functions during this time is to interfere with Pol III holoenzyme processivity. This creates a checkpoint, stops replication, and allows time to repair DNA lesions via the appropriate repair pathway.[13] Another function of Pol IV is to perform translesion synthesis at the stalled replication fork like, for example, bypassing N2-deoxyguanine adducts at a faster rate than transversing undamaged DNA. Cells lacking dinB gene have a higher rate of mutagenesis caused by DNA damaging agents.[14]
Pol V
DNA polymerase V (Pol V) is a Y-family DNA polymerase that is involved in SOS response and translesion synthesis DNA repair mechanisms.[15] Transcription of Pol V via the umuDC genes is highly regulated to produce only Pol V when damaged DNA is present in the cell generating an SOS repsonse. Stalled polymerases causes RecA to bind to the ssDNA, which causes the LexA protein to autodigest. LexA then loses is ability to repress the transcription of the umuDC operon. The same RecA-ssDNA nucleoprotein posttranslationally modifies the UmuD protein into UmuD' protein. UmuD and UmuD' form a heterodimer that interacts with UmuC, which in turn activates umuC's polymerase catalytic activity on damaged DNA.[16]
Eukaryotic DNA polymerase
Polymerases β, λ, σ and μ (Polymerases beta, lambda, sigma, and mu)
Family X polymerases contain the well-known eukaryotic polymerase pol β (beta), as well as other eukaryotic polymerases such as Pol σ (sigma), Pol λ (lambda), Pol μ (mu), and Terminal deoxynucleotidyl transferase (TdT). Family X polymerases are found mainly in vertebrates, and a few are found in plants and fungi. These polymerases have highly conserved regions that include two helix-hairpin-helix motifs that are imperative in the DNA-polymerase interactions. One motif is located in the 8 kDa domain that interacts with downstream DNA and one motif is located in the thumb domain that interacts with the primer strand. Pol β, encoded by POLB gene, is required for short-patch base excision repair, a DNA repair pathway that is essential for repairing alkylated or oxidized bases as well as abasic sites. Pol λ and Pol μ, encoded by the POLL and POLM genes respectively, are involved in non-homologous end-joining, a mechanism for rejoining DNA double-strand breaks due to hydrogen peroxide and ionizing radition, respectively. TdT is expressed only in lymphoid tissue, and adds "n nucleotides" to double-strand breaks formed during V(D)J recombination to promote immunological diversity.[17]
Polymerases α, δ and ε (Polymerases alpha, delta, and epsilon)
Pol α (alpha), Pol δ (delta), and Pol ε (epsilon) are members of Family B Polymerases and are the main polymerases involved with nuclear DNA replication. Pol α complex (pol α-DNA primase complex) consists of four subunits: the catalytic subunit POLA1, the regulatory subunit POLA2, and the small and the large primase subunits PRIM1 and PRIM2 respectively. Once primase has created the RNA primer, Pol α starts replication elongating the primer with ~20 nucleotides.[18] > Due to their high processivity, Pol ε and Pol δ take over the leading and lagging strand synthesis from Pol α respectively.[4]: 218–219 Pol δ is expressed by genes POLD1, creating the catalytic subunit, POLD2, POLD3, and POLD4 creating the other subunits that interact with Proliferating Cell Nuclear Antigen (PCNA), which is a DNA clamp that allows Pol δ to possess processivity.[19] Pol ε is encoded by the POLE, the catalytic subunit, POLE2, and POLE3 genes. While Pol ε's main function is to extend the leading strand during replication, Pol ε's C-terminus region is thought to be essential to cell vitality as well. The C-terminus region is thought to provide a checkpoint before entering anaphase, provide stability to the holoenzyme, and add proteins to the holoenzyme necessary for initiation of replication.[20]
Polymerases η, ι and κ (Polymerases eta, iota, and kappa)
Pol η (eta), Pol ι (iota), and Pol κ (kappa), are Family Y DNA polymerases involved in the DNA repair by translesion synthesis and encoded by genes POLH, POLI, and POLK respectively. Members of Family Y have five common motifs to aid in binding the substrate and primer terminus and they all include the typical right hand thumb, palm and finger domains with added domains like little finger (LF), polymerase-associated domain (PAD), or wrist. The active site, however, differs between family members due to the different lesions being repaired. Polymerases in Family Y are low-fidelity polymerases, but have been proven to do more good than harm as mutations that affect the polymerase can cause various diseases, such as skin cancer and Xeroderma Pigmentosum Variant (XPS). The importance of these polymerases is evidenced by the fact that gene encoding DNA polymerase η is referred as XPV, because loss of this gene results in the disease Xeroderma Pigmentosum Variant. Pol η is particularly important for allowing accurate translesion synthesis of DNA damage resulting from ultraviolet radiation. The functionality of Pol κ is not completely understood, but researchers have found two probable functions. Pol κ is thought to act as an extender or an inserter of a specific base at certain DNA lesions. All three translesion synthesis polymerases, along with Rev1, are recruited to damaged lesions via stalled replicative DNA polymerases. There are two pathways of damage repair leading researchers to conclude that the chosen pathway depends on which strand contains the damage, the leading or lagging strand.[21]
Polymerases Rev1 and ζ (Polymerases Rev1 and zeta)
Pol ζ another B family polymerase, is made of two subunits Rev3, the catalytic subunit, and Rev7, which increases the catalytic function of the polymerase, and is involved in translesion synthesis. Pol ζ lacks 3' to 5' exonuclease activity, is unique in that it can extend primers with terminal mismatches. Rev1 has three regions of interest in the BRCT domain, ubiquitin-binding domain, and C-terminal domain and has dCMP transferase ability, which adds deoxycytidine opposite lesions that would stall replicative polymerases Pol δ and Pol ε. These stalled polymerases activate ubiquitin complexes that in turn disassociate replication polymerases and recruit Pol ζ and Rev1. Together Pol ζ and Rev1 add deoxycytidine and Pol ζ extends past the lesion. Through a yet undetermined process, Pol ζ disassociates and replication polymerases reassociate and continue replication. Pol ζ and Rev1 are not required for replication, but loss of REV3 gene in budding yeast can cause increased sensitivity to DNA-damaging agents due to collapse of replication forks where replication polymerases have stalled.[22]
Telomerase
Telomerase is a ribonucleoprotein recruited to replicate ends of linear chromosomes because normal DNA polymerase cannot replicate the ends, or telomere. The single-strand 3’ overhang of the double-strand chromosome with the sequence 5’-TTAGGG-3’ recruits telomerase. Telomerase acts like other DNA polymerases by extending the 3’ end, but, unlike other DNA polymerases, telomerase does not require a template. The TERT subunit, an example of a reverse transcriptase, uses the RNA subunit to form the primer–template junction that allows telomerase to extend the 3’ end of chromosome ends. The gradual decrease in size of telomeres as the result of many replications over a lifetime are thought to be associated with the effects of aging.[4]: 248–249
Polymerases γ and θ (Polymerases gamma and theta)
Pol γ (gamma) and Pol θ (theta) are Family A polymerases. Pol γ, encoded by the POLG gene, is the only mtDNA polymerase and therefore replicates, repairs, and has proofreading 3'-5' exonuclease and 5' dRP lyase activities. Any mutation that leads to limited or non-functioning Pol γ has a significant effect on mtDNA and is the most common cause of autosomal inherited mitochondrial disorders.[23] Pol γ contains a C-terminus polymerase domain and an N-terminus 3'-5' exonuclease domain that are connected via the linker region, which binds the accessory subunit. The accessory subunit binds DNA and is required for processivity of Pol γ. Point mutation A467T in the linker region is responsible for more than one-third of all Pol γ-associated mitochondrial disorders.[24] While many homologs of Pol θ, encoded by the POLQ gene, are found in eukaryotes, its function is not clearly understood. The sequence of amino acids in the C-terminus is what classifies Pol θ as Family A polymerase, although the error rate for Pol θ is more closely related to Family Y polymerases. Pol θ extends mismatched primer termini and can bypass abasic sites by adding a nucleotide also has Deoxyribophosphodiesterase (dRPase) activity in the polymerase domain and can show ATPase activity in close proximity to ssDNA.[25]
Reverse transcriptase
Retroviruses encode an unusual DNA polymerase called reverse transcriptase, which is an RNA-dependent DNA polymerase (RdDp) that synthesizes DNA from a template of RNA. The reverse transcriptase family contain both DNA polymerase functionality and RNase H functionality, which degrades RNA base-paired to DNA. Some retrovirus examples include Hepatitis B virus and HIV.[4]
See also
- DNA repair
- DNA replication
- DNA sequencing
- Genetic recombination
- Molecular cloning
- Polymerase chain reaction
- Reverse transcription
- RNA polymerase
References
- ^ "The Nobel Prize in Physiology or Medicine 1959". Nobel Foundation. Retrieved December 1, 2012.
- ^ Tessman I, Kennedy MA (February 1994). "DNA polymerase II of Escherichia coli in the bypass of abasic sites in vivo". Genetics. 136 (2): 439–48. PMC 1205799. PMID 7908652.
- ^ Steitz TA (June 1999). "DNA polymerases: structural diversity and common mechanisms". J. Biol. Chem. 274 (25): 17395–8. doi:10.1074/jbc.274.25.17395. PMID 10364165.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d e Losick R, Watson JD, Baker TA, Bell S, Gann A, Levine MW (2008). Molecular biology of the gene (6th ed.). San Francisco: Pearson/Benjamin Cummings. ISBN 0-8053-9592-X.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ McCarthy D, Minner C, Bernstein H, Bernstein C (October 1976). "DNA elongation rates and growing point distributions of wild-type phage T4 and a DNA-delay amber mutant". J. Mol. Biol. 106 (4): 963–81. doi:10.1016/0022-2836(76)90346-6. PMID 789903.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Maga G, Hubscher U, Spadari S, Villani G (2010). DNA Polymerases: Discovery, Characterization and Functions in Cellular DNA Transactions. World Scientific Publishing Company. ISBN 981-4299-16-2.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Camps M, Loeb LA (February 2004). "When pol I goes into high gear: processive DNA synthesis by pol I in the cell". Cell Cycle. 3 (2): 116–8. PMID 14712068.
- ^ Banach-Orlowska M, Fijalkowska IJ, Schaaper RM, Jonczyk P (October 2005). "DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli". Mol. Microbiol. 58 (1): 61–70. doi:10.1111/j.1365-2958.2005.04805.x. PMID 16164549.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Banach-Orlowska M, Fijalkowska IJ, Schaaper RM, Jonczyk P (October 2005). "DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli". Mol. Microbiol. 58 (1): 61–70. doi:10.1111/j.1365-2958.2005.04805.x. PMID 16164549.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Olson MW, Dallmann HG, McHenry CS (December 1995). "DnaX complex of Escherichia coli DNA polymerase III holoenzyme. The chi psi complex functions by increasing the affinity of tau and gamma for delta.delta' to a physiologically relevant range". J. Biol. Chem. 270 (49): 29570–7. PMID 7494000.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "DNA Polymerase Families". News-medical.net. 2014-05-06. Retrieved 2014-06-28.
- ^ Goodman MF (2002). "Error-prone repair DNA polymerases in prokaryotes and eukaryotes". Annu. Rev. Biochem. 71: 17–50. doi:10.1146/annurev.biochem.71.083101.124707. PMID 12045089.
- ^ Mori T (2012). "Escherichia coli DinB inhibits replication fork progression without significantly inducing the SOS response". Genes Genet Syst. 87 (2): 75–87. doi:10.1266/ggs.87.75. PMID 22820381.
- ^ Jarosz DF (2007). "Proficient and accurate bypass of persistent DNA lesions by DinB DNA polymerases". Cell Cycle. 6 (7): 817–22. doi:10.4161/cc.6.7.4065. PMID 17377496.
- ^ Patel M, Jiang Q, Woodgate R, Cox MM, Goodman MF (June 2010). "A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V". Crit. Rev. Biochem. Mol. Biol. 45 (3): 171–84. doi:10.3109/10409238.2010.480968. PMC 2874081. PMID 20441441.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sutton MD, Walker GC (July 2001). "Managing DNA polymerases: coordinating DNA replication, DNA repair, and DNA recombination". Proc. Natl. Acad. Sci. U.S.A. 98 (15): 8342–9. doi:10.1073/pnas.111036998. PMC 37441. PMID 11459973.
- ^ Yamtich J, Sweasy JB (May 2010). "DNA polymerase family X: function, structure, and cellular roles". Biochim. Biophys. Acta. 1804 (5): 1136–50. doi:10.1016/j.bbapap.2009.07.008. PMC 2846199. PMID 19631767.
- ^ Chansky, Michael Lieberman, Allan Marks, Alisa Peet ; illustrations by Matthew (2012). Marks' basic medical biochemistry : a clinical approach (4th ed. ed.). Philadelphia: Wolter Kluwer Health/Lippincott Williams & Wilkins. p. chapter13. ISBN 160831572X.
{{cite book}}:|edition=has extra text (help)CS1 maint: multiple names: authors list (link) - ^ Chung DW, Zhang JA, Tan CK, Davie EW, So AG, Downey KM (December 1991). "Primary structure of the catalytic subunit of human DNA polymerase delta and chromosomal location of the gene". Proc. Natl. Acad. Sci. U.S.A. 88 (24): 11197–201. doi:10.1073/pnas.88.24.11197. PMC 53101. PMID 1722322.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Edwards S, Li CM, Levy DL, Brown J, Snow PM, Campbell JL (April 2003). "Saccharomyces cerevisiae DNA polymerase epsilon and polymerase sigma interact physically and functionally, suggesting a role for polymerase epsilon in sister chromatid cohesion". Mol. Cell. Biol. 23 (8): 2733–48. doi:10.1128/mcb.23.8.2733-2748.2003. PMC 152548. PMID 12665575.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ohmori H, Hanafusa T, Ohashi E, Vaziri C (2009). "Separate roles of structured and unstructured regions of Y-family DNA polymerases". Adv Protein Chem Struct Biol. 78: 99–146. doi:10.1016/S1876-1623(08)78004-0. PMC 3103052. PMID 20663485.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Gan GN, Wittschieben JP, Wittschieben BØ, Wood RD (January 2008). "DNA polymerase zeta (pol zeta) in higher eukaryotes". Cell Res. 18 (1): 174–83. doi:10.1038/cr.2007.117. PMID 18157155.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Zhang L, Chan SS, Wolff DJ (July 2011). "Mitochondrial disorders of DNA polymerase γ dysfunction: from anatomic to molecular pathology diagnosis". Arch. Pathol. Lab. Med. 135 (7): 925–34. doi:10.1043/2010-0356-RAR.1. PMC 3158670. PMID 21732785.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Stumpf JD, Copeland WC (January 2011). "Mitochondrial DNA replication and disease: insights from DNA polymerase γ mutations". Cell. Mol. Life Sci. 68 (2): 219–33. doi:10.1007/s00018-010-0530-4. PMC 3046768. PMID 20927567.
- ^ Hogg M, Sauer-Eriksson AE, Johansson E (March 2012). "Promiscuous DNA synthesis by human DNA polymerase θ". Nucleic Acids Res. 40 (6): 2611–22. doi:10.1093/nar/gkr1102. PMC 3315306. PMID 22135286.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
Further reading
- Burgers PM, Koonin EV, Bruford E, Blanco L, Burtis KC, Christman MF, Copeland WC, Friedberg EC, Hanaoka F, Hinkle DC, Lawrence CW, Nakanishi M, Ohmori H, Prakash L, Prakash S, Reynaud CA, Sugino A, Todo T, Wang Z, Weill JC, Woodgate R (November 2001). "Eukaryotic DNA polymerases: proposal for a revised nomenclature". J. Biol. Chem. 276 (47): 43487–90. doi:10.1074/jbc.R100056200. PMID 11579108.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link)
External links
- DNA+polymerases at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- PDB Molecule of the Month DNA polymerase
- Unusual repair mechanism in DNA polymerase lambda, Ohio State University, July 25, 2006.
- A great animation of DNA Polymerase from WEHI at 1:45 minutes in
- 3D macromolecular structures of DNA polymerase from the EM Data Bank(EMDB)
