User:IChindris/sandbox
TMEM261
Transmembrane protein 261 is a protein that in humans is encoded by the TMEM261 gene located on chromosome 9[1]. TMEM261 is also known as C9ORF123, Chromosome 9 Open Reading Frame 123 and Transmembrane Protein C9orf123[2].
An Error has occurred retrieving Wikidata item for infobox
Gene Features
TMEM261 is located at 9p24.1,its length is 91,891 base pairs (bp) on the reverse strand.[2]Its neighbouring gene is PTPRD located at 9p23-p24.3 also on the reverse strand and encodes protein tyrosine phosphatase receptor type delta.[1][2]
TMEM261 has a contains 2 exons and 1 intron, and 6 transcript variants, the largest consisting of 742bp with a protein 129 amino acids (aa) in length and 13,500 Daltons (Da) in size, and the smallest coding transcript variant being 381bp with a protein 69aa long and 6,100 Da in size.[3]
Protein Features
The coding variant of TMEM261 is a protein of 112aa with a molecular weight of 12,300 Da.[4] The isoelectric is predicted to be 10.2Cite error: The <ref> tag has too many names (see the help page)., with it's posttranslational modification is 9.9[5].
Structure
TMEM261 contains a domain of unknown function, DUF4536 (pfam15055), predicted as a helical membrane spanning domain about 45aa (47-92) in length with no known domain relationships.[6][7] Two further transmembrane helical domains are predicted of lengths 18aa (52-69) and 23aa (81-102)Cite error: The <ref> tag has too many names (see the help page)..There is also a low complexity region spanning 25aa (14-39).[8] The tertiary structure for TMEM261 has not yet been determined, however it's secondary structure is mostly composed of coiled-coils regions with beta strands and alpha helices in the transmembrane and domain of unknown function reigons. The N-terminal region of TMEM261 is composed of a disordered region[9] [10] which contains the low complexity region[8] which is not highly conserved amongst orthologues[11][12].
Modifications
A N-myristoylation domain is believed to be present in some TMEM261 protein variants.[5] Post-translational modifications include myristoylation of the N-terminal Glycine residue[5][13] of the TMEM261 protein as well as phosphorylation of Threonine 31.[14]
-
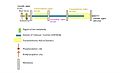
Annotated features of TMEM261 protein including topology and important sites for phosphorylation and Myristoylation as well DUF4536 and transmembrane helical domains.

Interactions
Proteins shown to interact with TMEM261 include NAAA, QTRT1,ZC4H2[15] and ZNF454[16][17]. It has also shown to interact with APP,ARHGEF38 and HNRNPD[18]. These proteins are involved with TMEM261 through a variety of interactions; protein-protein, RNA-protein and DNA-protein.
Expression
TMEM261 shows ubiquitous expression in humans detected in almost all tissue types in humans[19]. However, its expression is highest in the gall bladder, urinary bladder[20], pituitary gland, parathyroid and acites[19]. Staining intensity of cancer cells showed intermediate to high expression in breast, colorectal, ovarian, skin, urothelial head and neck cells. [20].TMEM261 and it's locus has been associated with colorectal cancer[21], breast cancer[22] and lymphomas[23][24] relating to gene amplification and rearrangements.
Evolution
Orthologues
TMEM261 shows conservation in vertebrates with higher overall conservation of its primary structure in mammals. High conservation in orthologues, including distant homologues, is seen from the domain of unknown function (DUF4536) to the C-terminus region, as the N-terminus reigon is only conserved highly in mammalian orthologues. The secondary structure of TMEM261 shows conservation across most orthologues.[11][25][26]
| Organism | Scientific Name | Accession Number | Date of Divergence from Humans (million years) | Amino acids (bp) | Identity (%) | Class |
|---|---|---|---|---|---|---|
| Humans | Homo sapiens | NP_219500.1 | 0 | 112 | 100 | Mammalia |
| Gorilla | Gorilla gorilla | XP_004047847.1 | 8.8 | 112 | 99 | Mammalia |
| Olive Baboon | Papio anubis | XP_003911767.1 | 29 | 112 | 84 | Mammalia |
| Sunda Flying Lemur | Galeopterus variegatus | XP_008587957.1 | 81.5 | 112 | 68 | Mammalia |
| Lesser Egyptian Jerboa | Jaculus Jaculus | XP_004653029.1 | 92.3 | 109 | 56 | Mammalia |
| Naked Mole Rat | Heterocephalus glaber | XP_004898193.1 | 92.3 | 114 | 45 | Mammalia |
| White Rhinoceros | Ceratotherium simum simum | XP_004436891.1 | 94.2 | 112 | 66 | Mammalia |
| Nine-banded armadillo | Dasypus novemcinctus | XP_004459147.1 | 104.4 | 112 | 59 | Mammalia |
| Green Sea Turtle | Chelonia mydas | XP_007056940.1 | 296 | 85 | 49 | Reptilia |
| Zebra Finch | Taeniopygia Guttata | XP_002187613.2 | 296 | 72 | 47 | Aves |
| Western Clawed Frog | Xenopus tropicalis | XP_002943025.1 | 371.2 | 85 | 45 | Amphibia |
| Haplochromis burtoni | Haplochromis burtoni | XP_005928614.1 | 400.1 | 91 | 51 | Actinopterygii |
| Australian Ghost Shark | Callorhinchus milii | XP_007884223.1 | 426.5 | 86 | 43 | Chondrichthyes |
Paralogues
Tmem261 has no known paralogs. It has orthologs and paralogs limited to vertebrates, it's oldest homolog dating to that of the cartilaginous fishes[26] which diverged from Homo sapiens 462.5 million years ago. [27]
References
- ^ a b "Entrez Protein: TMEM261".
- ^ a b c "GeneCards:TMEM261 Gene". Cite error: The named reference "GeneCards" was defined multiple times with different content (see the help page).
- ^ Thierry-Mieg, D; Thierry-Mieg, J. (2006). "AceView: a comprehensive cDNA-supported gene and transcripts annotation". Genome Biology. 7 (Suppl 1): S12. doi:10.1186/gb-2006-7-s1-s12. PMC 1810549. PMID 16925834.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ "Ensemble:Transcript TMEM261-003".
- ^ a b c "AceView:Homo sapiens gene C9orf123".
- ^ "NCBI Conserved Domains: DUF4536".
- ^ "EMBL-EBI Interpro: Transmembrane protein 261 (Q96GE9)".
- ^ a b "Vega: Transcript: C9orf123-003".
- ^ "Protein structure prediction on the web: a case study using the Phyre server". PHYRE: Protein Homology/analogY Recognition Engin.
- ^ Kelley, LA (2009). "Stenberg". MJE. 4: 363–371. doi:10.1038/nprot.2009.2. PMID 19247286.
- ^ a b "ClustalW".
- ^ Thompson, Julie D; Higgins, Desmond G; Gibson, Toby J (1994). "CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice". Nucleic Acids Res. 22 (22): 4673–4680. PMID 308517.
- ^ Gallo, Vincenzo. "Myristoylation : Proteins Post-translational Modifications". http://flipper.diff.org/. University of Turin.
{{cite web}}: External link in|website= - ^ "Nextprot:TMEM261 » Transmembrane protein 261".
- ^ Dash, A; et al. (2002). "Changes in differential gene expression because of warm ischemia time of radical prostatectomy specimens". Am J Pathol. 161 (5): 1743–1748. doi:10.1016/S0002-9440(10)64451-3.
{{cite journal}}: Explicit use of et al. in:|first1=(help) - ^ Rovillain, E; et al. (2011). "An RNA interference screen for identifying downstream effectors of the p53 and pRB tumour suppressor pathways involved in senescence". BMC Genomics. 12 (355). doi:10.1186/1471-2164-12-355. PMID 21740549.
{{cite journal}}: Explicit use of et al. in:|first1=(help)CS1 maint: unflagged free DOI (link) - ^ "c9orf123 protein (Homo Sapiens)- STRING Network View". STRING - Known and Predicted Protein-Protein Interactions.
- ^ "9ORF123 chromosome 9 open reading frame 123". BioGRID: Database of Protein and Genetic Interactions. TyersLab.
- ^ a b "EST profile: TMEM261". UniGene. National Library of Medicine.
- ^ a b "The Human Protein Atlas:TMEM261".
- ^ Gaspar, C (2008). "Cross-Species Comparison of Human and Mouse Intestinal Polyps Reveals Conserved Mechanisms in Adenomatous Polyposis Coli (APC)-Driven Tumorigenesis". Am J Pathol. 172 (5): 1363–1380. doi:10.2353/ajpath.2008.070851. PMID 18403596.
- ^ Wu, J (2012). "Identification and functional analysis of 9p24 amplified genes in human breast cancer". Oncogene. 31 (3): 333–341. doi:10.1038/onc.2011.227. PMID 21666724.
- ^ Twa, D D W; et al. (2014). "Genomic Rearrangements Involving Programmed Death Ligands Are Recurrent in Primary Mediastinal Large B-Cell Lymphoma". Blood. 123 (13): 2062–2065. doi:10.1182/blood-2013-10-535443. PMID 24497532.
{{cite journal}}: Explicit use of et al. in:|first1=(help) - ^ Green, M R; et al. (2010). "Integrative Analysis Reveals Selective 9p24.1 Amplification, Increased PD-1 Ligand Expression, and Further Induction via JAK2 in Nodular Sclerosing Hodgkin Lymphoma and Primary Mediastinal Large B-Cell Lymphoma". Blood. 116 (17): 3268–3277. doi:10.1182/blood-2010-05-282780. PMID 20628145.
{{cite journal}}: Explicit use of et al. in:|first1=(help) - ^ Thompson, Julie D; Higgins, Desmond G; Gibson, Toby J (1994). "CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice". Nucleic Acids Res. 22 (22): 4673–4680. PMID 308517.
- ^ a b "NCBI BLAST:Basic Local Alignment Search Tool".
- ^ Hedges, S. Blaire; Dudley, Joel; Kumar, Sudhir (22 September 2006). "TimeTree: a public knowledge-base of divergence times among organisms" (PDF). 22 (23): 2971–2972. doi:10.1093/bioinformatics/btl505.
{{cite journal}}: Cite has empty unknown parameter:|1=(help); Cite journal requires|journal=(help)
External Links
- PubMed
- NCBI gene record
- GeneCards
- UCSC Genome Browser
- Expasy Bioinformatics Resource Portal
- SDSC Biology Workbench
Further Reading
- Nicholas K. Tonks (2006). "Protein tyrosine phosphatases: from genes, to function, to disease". Cancer Cell. 7: 833–846. doi:10.1038/nrm2039.
{{cite journal}}: Unknown parameter|author-separator=ignored (help) - Merryweather-Clarke AT; et al. (2011). "Global gene expression analysis of human erythroid progenitors". Blood. 117 (13): e96-108. doi:10.1182/blood-2010-07-290825. PMID 21270440.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|author-separator=ignored (help) - Welch JJ, Watts JA, Vakoc CR; et al. (2004). "Global regulation of erythroid gene expression by transcription factor GATA-1". Blood. 104 (10): 3136–3147. doi:10.1182/blood-2004-04-1603. PMID 15297311.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|author-separator=ignored (help)CS1 maint: multiple names: authors list (link) - Nickeleit I; et al. (2008). "Argyrin a reveals a critical role for the tumor suppressor protein p27(kip1) in mediating antitumor activities in response to proteasome inhibition". Cancer Cell. 14 (1): 23–35. doi:10.1016/j.ccr.2008.05.016. PMID 18598941.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|author-separator=ignored (help)