Tissue transglutaminase
| Protein-glutamine gamma-glutamyltransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 2.3.2.13 | ||||||||
| CAS no. | 80146-85-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Tissue transglutaminase (abbreviated as tTG or TG2) is a 78-kDa, calcium-dependent enzyme (EC 2.3.2.13) of the protein-glutamine γ-glutamyltransferases family (or simply transglutaminase family).[5][6] Like other transglutaminases, it crosslinks proteins between an ε-amino group of a lysine residue and a γ-carboxamide group of glutamine residue, creating an inter- or intramolecular bond that is highly resistant to proteolysis (protein degradation). Aside from its crosslinking function, tTG catalyzes other types of reactions including deamidation, GTP-binding/hydrolyzing, and isopeptidase activities.[7] Unlike other members of the transglutaminase family, tTG can be found both in the intracellular and the extracellular spaces of various types of tissues and is found in many different organs including the heart, the liver, and the small intestine. Intracellular tTG is abundant in the cytosol but smaller amounts can also be found in the nucleus and the mitochondria.[6] Intracellular tTG is thought to play an important role in apoptosis.[8] In the extracellular space, tTG binds to proteins of the extracellular matrix (ECM),[9] binding particularly tightly to fibronectin.[10] Extracellular tTG has been linked to cell adhesion, ECM stabilization, wound healing, receptor signaling, cellular proliferation, and cellular motility.[6]
tTG is particularly notable for being the autoantigen in celiac disease, a lifelong illness in which the consumption of dietary gluten causes a pathological immune response resulting in the inflammation of the small intestine and subsequent villous atrophy.[11][12][13]
Structure
Gene
The human tTG gene is located on the 20th chromosome (20q11.2-q12).
Protein
TG2 is a multifunctional enzyme that belongs to transglutaminases which catalyze the crosslinking of proteins by epsilon-(gamma-glutamyl)lysine isopeptide bonds.[14] Crystal structures of TG2 with bound GDP, GTP, or ATP have demonstrated that these forms of TG2 adopt a "closed" conformation, whereas TG2 with the active site occupied by an inhibitory gluten peptide mimic or other similar inhibitors adopts an "open" conformation. In the open conformation the four domains of TG2 are arranged in an extended configuration, whereas in the closed conformation the two C-terminal domains are folded in on the catalytic core domain. The N-terminal domain only shows minor structural changes between the two different conformations.[15]
Mechanism
The catalytic mechanism for crosslinking in human tTG involves the thiol group from a Cys residue in the active site of tTG.[6] The thiol group attacks the carboxamide of a glutamine residue on the surface of a protein or peptide substrate, releasing ammonia, and producing a thioester intermediate. The thioester intermediate can then be attacked by the surface amine of a second substrate (typically from a lysine residue). The end product of the reaction is a stable isopeptide bond between the two substrates (i.e. crosslinking). Alternatively, the thioester intermediate can be hydrolyzed, resulting in the net conversion of the glutamine residue to glutamic acid (i.e. deamidation).[6] The deamidation of glutamine residues catalyzed by tTG is thought to be linked to the pathological immune response to gluten in celiac disease.[12] A schematic for the crosslinking and the deamidation reactions is provided in Figure 1.

Regulation
Crosslinking activity by tTG requires the binding of Ca2+ ions.[16] Multiple Ca2+ can bind to a single tTG molecule.[6] In contrast, the binding of one molecule of GTP or GDP inhibits the crosslinking activity of the enzyme.[16] Therefore, intracellular tTG is mostly inactive due to the relatively high concentration of GTP/GDP and the low levels of calcium inside the cell.[6][12] Although extracellular tTG is expected to be active due to the low concentration of guanine nucleotides and the high levels of calcium in the extracellular space, evidence has shown that extracellular tTG is mostly inactive.[6][12][16] Recent studies suggest that extracellular tTG is kept inactive by the formation of a disulfide bond between two vicinal Cys residues. Therefore, oxidation/reduction of the disulfide bond serves as a third allosteric regulatory mechanism (along with GTP/GDP and Ca2+) for the activation of tTG.[12] Thioredoxin has been shown to activate extracellular tTG by reducing the disulfide bond.[16] Recent studies have suggested that interferon-γ may serve as an activator of extracellular tTG in the small intestine; these studies have a direct implication to the pathogenesis of celiac disease.[12] Activation of tTG has been shown to be accompanied by large conformational changes, switching from a compact (inactive) to an extended (active) conformation. (see Figure 2)[16][17][18]

Function
tTG is expressed ubiquitously. It requires calcium as a cofactor for transamidation activity. Transcription is increased by retinoic acid. Among its many supposed functions, it appears to play a role in wound healing, apoptosis, and extracellular matrix development[11]
tTG is thought to be involved in the regulation of the cytoskeleton by crosslinking various cytoskeletal proteins including myosin, actin, and spectrin.[19] Evidence shows that intracellular tTG crosslinks itself to myosin. It is also believed that tTG may stabilize the structure of the dying cells during apoptosis by polymerizing the components of the cytoskeleton, therefore preventing the leakage of the cellular contents into the extracellular space.[7]
tTG also has GTPase activity:[5] In the presence of GTP, it suggested to function as a G protein participating in signaling processes.[20] Besides its transglutaminase activity, tTG is proposed to also act as kinase,[21] and protein disulfide isomerase,[22] and deamidase.[23] This latter activity is important in the deamidation of gliadin peptides, thus playing important role in the pathology of coeliac disease.
Clinical significance
tTG is best known for its link with celiac disease.[13] Anti-transglutaminase antibodies result in a form of gluten sensitivity in which a cellular response to Triticeae glutens that are crosslinked to tTG are able to stimulate transglutaminase specific B-cell responses that eventually result in the production of anti-transglutaminase antibodies IgA and IgG.[24]
tTG is believed to be involved in several neurodegenerative disorders including Alzheimer, Parkinson and Huntington diseases.[25][26] Such neurological diseases are characterized in part by the abnormal aggregation of proteins due to the increased activity of protein crosslinking in the affected brain.[27] Additionally, specific proteins associated with these disorders have been found to be in vivo and in vitro substrates of tTG.[7] Although tTG is up regulated in the areas of the brain affected by Huntington's disease, a recent study showed that increasing levels of tTG do not affect the onset and/or progression of the disease in mice.[28]
Recent studies suggest that tTG also plays a role in inflammation and tumor biology.[11] tTG expression is elevated in multiple cancer cell types and is implicated in drug resistance and metastasis due to its ability to promote mesenchymal transition and stem cell like properties.
Diagnostic
Serology for anti-tTG antibodies has superseded older serological tests (anti-endomysium, anti-gliadin, and anti-reticulin) and has a strong sensitivity (99%) and specificity (>90%) for identifying celiac disease. Modern anti-tTG assays rely on a human recombinant protein as an antigen.[29]
Therapeutic
It's still experimental to use tTG as a form of surgical glue. It is also being studied as an attenuator of metastasis in certain tumors.[11]
Interactions
TG2 participates in both enzymatic and non-enzymatic interactions. Enzymatic interactions are formed between TG2 and its substrate proteins containing the glutamine donor and lysine donor groups in the presence of calcium. Substrates of TG2 are known to affect TG2 activity, which enables it to subsequently execute diverse biological functions in the cell. However, the importance of non-enzymatic interactions in regulating TG2 activities is yet to be revealed. Recent studies indicate that non-enzymatic interactions play physiological roles and enable diverse TG2 functions in a context-specific manner.[30]
| Mouse Mutant Alleles for Tgm2 | |
|---|---|
| Marker Symbol for Mouse Gene. This symbol is assigned to the genomic locus by the MGI | Tgm2 |
| Mutant Mouse Embryonic Stem Cell Clones. These are the known targeted mutations for this gene in a mouse. | Tgm2tm1a(KOMP)Wtsi |
| Example structure of targeted conditional mutant allele for this gene | |
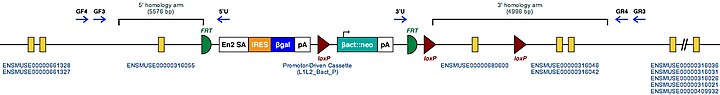
| |
| These Mutant ES Cells can be studied directly or used to generate mice with this gene knocked out. Study of these mice can shed light on the function of Tgm2: see Knockout mouse | |
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000198959 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000037820 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b Király R, Demény M, Fésüs L (Dec 2011). "Protein transamidation by transglutaminase 2 in cells: a disputed Ca2+-dependent action of a multifunctional protein". The FEBS Journal. 278 (24): 4717–39. doi:10.1111/j.1742-4658.2011.08345.x. PMID 21902809.
- ^ a b c d e f g h Klöck C, Diraimondo TR, Khosla C (Jul 2012). "Role of transglutaminase 2 in celiac disease pathogenesis". Seminars in Immunopathology. 34 (4): 513–22. doi:10.1007/s00281-012-0305-0. PMC 3712867. PMID 22437759.
- ^ a b c Facchiano F, Facchiano A, Facchiano AM (2006). "The role of transglutaminase-2 and its substrates in human diseases". Frontiers in Bioscience. 11: 1758–73. doi:10.2741/1921. PMID 16368554.
- ^ McConkey DJ, Orrenius S (Oct 1997). "The role of calcium in the regulation of apoptosis". Biochemical and Biophysical Research Communications. 239 (2): 357–66. doi:10.1006/bbrc.1997.7409. PMID 9344835.
- ^ Lortat-Jacob H, Burhan I, Scarpellini A, Thomas A, Imberty A, Vivès RR, Johnson T, Gutierrez A, Verderio EA (May 2012). "Transglutaminase-2 interaction with heparin: identification of a heparin binding site that regulates cell adhesion to fibronectin-transglutaminase-2 matrix". The Journal of Biological Chemistry. 287 (22): 18005–17. doi:10.1074/jbc.M111.337089. PMC 3365763. PMID 22442151.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Akimov SS, Krylov D, Fleischman LF, Belkin AM (Feb 2000). "Tissue transglutaminase is an integrin-binding adhesion coreceptor for fibronectin". The Journal of Cell Biology. 148 (4): 825–38. doi:10.1083/jcb.148.4.825. PMC 2169362. PMID 10684262.
- ^ a b c d Griffin M, Casadio R, Bergamini CM (Dec 2002). "Transglutaminases: nature's biological glues". The Biochemical Journal. 368 (Pt 2): 377–96. doi:10.1042/BJ20021234. PMC 1223021. PMID 12366374.
- ^ a b c d e f Diraimondo TR, Klöck C, Khosla C (Apr 2012). "Interferon-γ activates transglutaminase 2 via a phosphatidylinositol-3-kinase-dependent pathway: implications for celiac sprue therapy". The Journal of Pharmacology and Experimental Therapeutics. 341 (1): 104–14. doi:10.1124/jpet.111.187385. PMC 3310700. PMID 22228808.
- ^ a b Di Sabatino A, Vanoli A, Giuffrida P, Luinetti O, Solcia E, Corazza GR (Aug 2012). "The function of tissue transglutaminase in celiac disease". Autoimmunity Reviews. 11 (10): 746–53. doi:10.1016/j.autrev.2012.01.007. PMID 22326684.
- ^ "Entrez Gene: TGM2 transglutaminase 2".
- ^ Chen X, Hnida K, Graewert MA, Andersen JT, Iversen R, Tuukkanen A, Svergun D, Sollid LM (Aug 2015). "Structural Basis for Antigen Recognition by Transglutaminase 2-specific Autoantibodies in Celiac Disease". The Journal of Biological Chemistry. 290 (35): 21365–75. doi:10.1074/jbc.M115.669895. PMC 4571865. PMID 26160175.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d e Jin X, Stamnaes J, Klöck C, DiRaimondo TR, Sollid LM, Khosla C (Oct 2011). "Activation of extracellular transglutaminase 2 by thioredoxin". The Journal of Biological Chemistry. 286 (43): 37866–73. doi:10.1074/jbc.M111.287490. PMC 3199528. PMID 21908620.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Pinkas DM, Strop P, Brunger AT, Khosla C (Dec 2007). "Transglutaminase 2 undergoes a large conformational change upon activation". PLoS Biology. 5 (12): e327. doi:10.1371/journal.pbio.0050327. PMC 2140088. PMID 18092889.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Colak G, Keillor JW, Johnson GV (2011). Polymenis M (ed.). "Cytosolic guanine nucledotide binding deficient form of transglutaminase 2 (R580a) potentiates cell death in oxygen glucose deprivation". PLOS ONE. 6 (1): e16665. doi:10.1371/journal.pone.0016665. PMC 3031627. PMID 21304968.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Nurminskaya MV, Belkin AM (2012). "Cellular functions of tissue transglutaminase". International Review of Cell and Molecular Biology. International Review of Cell and Molecular Biology. 294: 1–97. doi:10.1016/B978-0-12-394305-7.00001-X. ISBN 9780123943057. PMC 3746560. PMID 22364871.
- ^ Fesus L, Piacentini M (Oct 2002). "Transglutaminase 2: an enigmatic enzyme with diverse functions". Trends in Biochemical Sciences. 27 (10): 534–9. doi:10.1016/S0968-0004(02)02182-5. PMID 12368090.
- ^ Mishra S, Murphy LJ (Jun 2004). "Tissue transglutaminase has intrinsic kinase activity: identification of transglutaminase 2 as an insulin-like growth factor-binding protein-3 kinase". The Journal of Biological Chemistry. 279 (23): 23863–8. doi:10.1074/jbc.M311919200. PMID 15069073.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Hasegawa G, Suwa M, Ichikawa Y, Ohtsuka T, Kumagai S, Kikuchi M, Sato Y, Saito Y (Aug 2003). "A novel function of tissue-type transglutaminase: protein disulphide isomerase". The Biochemical Journal. 373 (Pt 3): 793–803. doi:10.1042/BJ20021084. PMC 1223550. PMID 12737632.
- ^ Sakly W, Thomas V, Quash G, El Alaoui S (Dec 2006). "A role for tissue transglutaminase in alpha-gliadin peptide cytotoxicity". Clinical and Experimental Immunology. 146 (3): 550–8. doi:10.1111/j.1365-2249.2006.03236.x. PMC 1810403. PMID 17100777.
- ^ Dieterich W, Ehnis T, Bauer M, Donner P, Volta U, Riecken EO, Schuppan D (Jul 1997). "Identification of tissue transglutaminase as the autoantigen of celiac disease". Nature Medicine. 3 (7): 797–801. doi:10.1038/nm0797-797. PMID 9212111.
- ^ Wilhelmus MM, Verhaar R, Andringa G, Bol JG, Cras P, Shan L, Hoozemans JJ, Drukarch B (Mar 2011). "Presence of tissue transglutaminase in granular endoplasmic reticulum is characteristic of melanized neurons in Parkinson's disease brain". Brain Pathology. 21 (2): 130–9. doi:10.1111/j.1750-3639.2010.00429.x. PMID 20731657.
- ^ Ricotta M, Iannuzzi M, Vivo GD, Gentile V (May 2010). "Physio-pathological roles of transglutaminase-catalyzed reactions". World Journal of Biological Chemistry. 1 (5): 181–7. doi:10.4331/wjbc.v1.i5.181. PMC 3083958. PMID 21541002.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Martin A, Giuliano A, Collaro D, De Vivo G, Sedia C, Serretiello E, Gentile V (Jan 2013). "Possible involvement of transglutaminase-catalyzed reactions in the physiopathology of neurodegenerative diseases". Amino Acids. 44 (1): 111–8. doi:10.1007/s00726-011-1081-1. PMID 21938398.
- ^ Kumar A, Kneynsberg A, Tucholski J, Perry G, van Groen T, Detloff PJ, Lesort M (Sep 2012). "Tissue transglutaminase overexpression does not modify the disease phenotype of the R6/2 mouse model of Huntington's disease". Experimental Neurology. 237 (1): 78–89. doi:10.1016/j.expneurol.2012.05.015. PMC 3418489. PMID 22698685.
- ^ Sblattero D, Berti I, Trevisiol C, Marzari R, Tommasini A, Bradbury A, Fasano A, Ventura A, Not T (May 2000). "Human recombinant tissue transglutaminase ELISA: an innovative diagnostic assay for celiac disease". The American Journal of Gastroenterology. 95 (5): 1253–7. doi:10.1111/j.1572-0241.2000.02018.x. PMID 10811336.
- ^ Kanchan K, Fuxreiter M, Fésüs L (Aug 2015). "Physiological, pathological, and structural implications of non-enzymatic protein-protein interactions of the multifunctional human transglutaminase 2". Cellular and Molecular Life Sciences. 72 (16): 3009–35. doi:10.1007/s00018-015-1909-z. PMID 25943306.
External links
- Endomysial antibodies
- A collection of substrates and interaction partners of TG2 is accessible in the TRANSDAB, an interactive transglutaminase substrate database.