ATAC-seq
This article has multiple issues. Please help improve it or discuss these issues on the talk page. (Learn how and when to remove these messages)
|
ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) is a technique used in molecular biology to assess genome-wide chromatin accessibility.[1] In 2013, the technique was first described as an alternative advanced method for MNase-seq (sequencing of micrococcal nuclease sensitive sites), FAIRE-Seq and DNase-Seq.[1] ATAC-seq is a faster and more sensitive analysis of the epigenome than DNase-seq or MNase-seq.[2][3][4]
Description
ATAC-seq identifies accessible DNA regions by probing open chromatin with hyperactive mutant Tn5 Transposase that inserts sequencing adapters into open regions of the genome. [2][5] While naturally occurring transposases have a low level of activity, ATAC-seq employs the mutated hyperactive transposase.[6] In a process called "tagmentation", Tn5 transposase cleaves and tags double-stranded DNA with sequencing adaptors.[7] The tagged DNA fragments are then purified, PCR-amplified, and sequenced using next-generation sequencing.[7] Sequencing reads can then be used to infer regions of increased accessibility as well as to map regions of transcription factor binding sites and nucleosome positions.[2] The number of reads for a region correlate with how open that chromatin is, at single nucleotide resolution.[2] ATAC-seq requires no sonication or phenol-chloroform extraction like FAIRE-seq;[8] no antibodies like ChIP-seq;[9] and no sensitive enzymatic digestion like MNase-seq or DNase-seq.[10] ATAC-seq preparation can be completed in under three hours.[11]
Applications
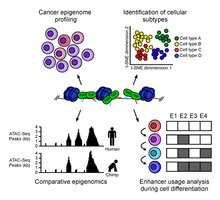
ATAC-Seq analysis is used to investigate a number of chromatin-accessibility signatures. The most common use is nucleosome mapping experiments,[3] but it can be applied to mapping transcription factor biding sites,[12] adapted to map DNA methylation sites,[13] or combined with sequencing techniques.[14]
The utility of high-resolution enhancer mapping ranges from studying the evolutionary divergence of enhancer usage (e.g. between chimps and humans) during development[15] and uncovering a lineage-specific enhancer map used during blood cell differentiation.[16]
ATAC-Seq has also been applied to defining the genome-wide chromatin accessibility landscape in human cancers,[17] and revealing an overall decrease in chromatin accessibility in macular degeneration.[18] Computational footprinting methods can be performed on ATAC-seq to find cells specific binding sites and transcription factors with cell specific activity.[19]
Single-cell ATAC-seq
Modifications to the ATAC-seq protocol have been made to accommodate single-cell analysis. Microfluidics can be used to separate single nuclei and perform ATAC-seq reactions individually.[11] With this approach, single cells are captured by either a microfluidic device or a liquid deposition system before tagmentation.[11][20] An alternative technique that does not require single cell isolation is combinatorial cellular indexing. This technique uses barcoding to measure chromatin accessibility in thousands of individual cells; it can generate epigenomic profiles from 10,000-100,000 cells per experiment.[21] But combinatorial cellular indexing requires additional, custom-engineered equipment or a large quantity of custom, modified Tn5.[22]
Efficiency and specifications
Methods to assess open chromatin via next generation sequencing (NGS) have been in use for a number of years. The main advantage of ATAC-seq over existing methods is the simplicity of the library preparation protocol: Tn5 insertion followed by two rounds of PCR. After library preparation, the DNA is sequenced with NGS technology, and the number of reads for a region correlate with how open that chromatin is at a single nucleotide resolution. ATAC-seq requires no sonication or phenol-chloroform extraction like FAIRE-seq; no antibodies like ChIP-seq; and no sensitive enzymatic digestion like MNase-seq or DNase-seq. Unlike similar methods, which can take up to four days to complete, ATAC-seq preparation can be completed in under three hours.[11]
Another consideration in the ATAC-seq protocol is cell number. While the total number of cells defines library complexity, too few cells leads to under-transposition, while too many leads to over-transposition. It is therefore important to optimize cell number from the beginning: for human studies, 500–50,000 cells are recommended, but this can vary between species and cell type.
The advantages of ATAC-seq include:
- Low requirements on the amount of the biological sample. 50,000 cells are sufficient for this technique, as opposed to others like MNase-seq or DNase-seq that require at least 1,000-fold more material.[1][2]
- Speed: The whole protocol requires 3 hours in total.[1][2][3]
References
- ^ a b c d Buenrostro, Jason D; Giresi, Paul G; Zaba, Lisa C; Chang, Howard Y; Greenleaf, William J (2013). "Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position". Nature Methods. 10 (12): 1213–1218. doi:10.1038/nmeth.2688. ISSN 1548-7091. PMC 3959825.
- ^ a b c d e f Buenrostro, Jason D.; Wu, Beijing; Chang, Howard Y.; Greenleaf, William J. (2015). "ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide": 21.29.1–21.29.9. doi:10.1002/0471142727.mb2129s109.
{{cite journal}}: Cite journal requires|journal=(help) - ^ a b c Schep, Alicia N.; Buenrostro, Jason D.; Denny, Sarah K.; Schwartz, Katja; Sherlock, Gavin; Greenleaf, William J. (2015). "Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions". Genome Research. 25 (11): 1757–1770. doi:10.1101/gr.192294.115. ISSN 1088-9051.
- ^ Song, L.; Crawford, G. E. (2010). "DNase-seq: A High-Resolution Technique for Mapping Active Gene Regulatory Elements across the Genome from Mammalian Cells". Cold Spring Harbor Protocols. 2010 (2): pdb.prot5384–pdb.prot5384. doi:10.1101/pdb.prot5384. ISSN 1559-6095.
- ^ Bajic, Marko; Maher, Kelsey A.; Deal, Roger B. (2018). "Identification of Open Chromatin Regions in Plant Genomes Using ATAC-Seq". 1675: 183–201. doi:10.1007/978-1-4939-7318-7_12. ISSN 1064-3745.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Reznikoff, William S. (2008). "Transposon Tn5". Annual Review of Genetics. 42 (1): 269–286. doi:10.1146/annurev.genet.42.110807.091656. ISSN 0066-4197.
- ^ a b Picelli, Simone; Björklund, Åsa K.; Reinius, Björn; Sagasser, Sven; Winberg, Gösta; Sandberg, Rickard (2014). "Tn5 transposase and tagmentation procedures for massively scaled sequencing projects". Genome Research. 24 (12): 2033–2040. doi:10.1101/gr.177881.114. ISSN 1088-9051.
- ^ Simon, Jeremy M; Giresi, Paul G; Davis, Ian J; Lieb, Jason D (2012). "Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA". Nature Protocols. 7 (2): 256–267. doi:10.1038/nprot.2011.444. ISSN 1754-2189.
- ^ Savic, Daniel; Partridge, E. Christopher; Newberry, Kimberly M.; Smith, Sophia B.; Meadows, Sarah K.; Roberts, BrianS.; Mackiewicz, Mark; Mendenhall, Eric M.; Myers, Richard M. (2015). "CETCh-seq: CRISPR epitope tagging ChIP-seq of DNA-binding proteins". Genome Research. 25 (10): 1581–1589. doi:10.1101/gr.193540.115. ISSN 1088-9051.
- ^ Hoeijmakers, Wieteke Anna Maria; Bártfai, Richárd (2018). "Characterization of the Nucleosome Landscape by Micrococcal Nuclease-Sequencing (MNase-seq)". 1689: 83–101. doi:10.1007/978-1-4939-7380-4_8. ISSN 1064-3745.
{{cite journal}}: Cite journal requires|journal=(help) - ^ a b c d Buenrostro, Jason D.; Wu, Beijing; Litzenburger, Ulrike M.; Ruff, Dave; Gonzales, Michael L.; Snyder, Michael P.; Chang, Howard Y.; Greenleaf, William J. (2015). "Single-cell chromatin accessibility reveals principles of regulatory variation". Nature. 523 (7561): 486–490. doi:10.1038/nature14590. ISSN 0028-0836.
- ^ Li, Zhijian; Schulz, Marcel H.; Zenke, Martin; Costa, Ivan G. (2018-07-08). "Identification of Transcription Factor Binding Sites using ATAC-seq". dx.doi.org. Retrieved 2019-09-09.
- ^ Soloway, Paul D.; Mimoso, Claudia A.; Tippens, Nathaniel D.; Spektor, Roman (2019-06-01). "methyl-ATAC-seq measures DNA methylation at accessible chromatin". Genome Research. 29 (6): 969–977. doi:10.1101/gr.245399.118. ISSN 1088-9051. PMID 31160376.
- ^ Hendrickson, David G.; Soifer, Ilya; Wranik, Bernd J.; Botstein, David; Scott McIsaac, R. (2018), "Simultaneous Profiling of DNA Accessibility and Gene Expression Dynamics with ATAC-Seq and RNA-Seq", Methods in Molecular Biology, Springer New York, pp. 317–333, ISBN 9781493986170, retrieved 2019-09-09
- ^ Prescott, Sara L.; Srinivasan, Rajini; Marchetto, Maria Carolina; Grishina, Irina; Narvaiza, Iñigo; Selleri, Licia; Gage, Fred H.; Swigut, Tomek; Wysocka, Joanna (2015). "Enhancer Divergence and cis-Regulatory Evolution in the Human and Chimp Neural Crest". Cell. 163 (1): 68–83. doi:10.1016/j.cell.2015.08.036. ISSN 0092-8674.
{{cite journal}}: no-break space character in|first1=at position 5 (help); no-break space character in|first3=at position 6 (help); no-break space character in|first7=at position 5 (help) - ^ Lara-Astiaso, D.; Weiner, A.; Lorenzo-Vivas, E.; Zaretsky, I.; Jaitin, D. A.; David, E.; Keren-Shaul, H.; Mildner, A.; Winter, D.; Jung, S.; Friedman, N.; Amit, I. (2014). "Chromatin state dynamics during blood formation". Science. 345 (6199): 943–949. doi:10.1126/science.1256271. ISSN 0036-8075.
- ^ Corces, M. Ryan; Granja, Jeffrey M.; Shams, Shadi; Louie, Bryan H.; Seoane, Jose A.; Zhou, Wanding; Silva, Tiago C.; Groeneveld, Clarice; Wong, Christopher K.; Cho, Seung Woo; Satpathy, Ansuman T.; Mumbach, Maxwell R.; Hoadley, Katherine A.; Robertson, A. Gordon; Sheffield, Nathan C.; Felau, Ina; Castro, Mauro A. A.; Berman, Benjamin P.; Staudt, Louis M.; Zenklusen, Jean C.; Laird, Peter W.; Curtis, Christina; Greenleaf, William J.; Chang, Howard Y. (2018). "The chromatin accessibility landscape of primary human cancers". Science. 362 (6413): eaav1898. doi:10.1126/science.aav1898. ISSN 0036-8075.
- ^ Wang, Jie; Zibetti, Cristina; Shang, Peng; Sripathi, Srinivasa R.; Zhang, Pingwu; Cano, Marisol; Hoang, Thanh; Xia, Shuli; Ji, Hongkai; Merbs, Shannath L.; Zack, Donald J.; Handa, James T.; Sinha, Debasish; Blackshaw, Seth; Qian, Jiang (2018). "ATAC-Seq analysis reveals a widespread decrease of chromatin accessibility in age-related macular degeneration". Nature Communications. 9 (1). doi:10.1038/s41467-018-03856-y. ISSN 2041-1723.
- ^ Li, Zhijian; Schulz, Marcel H.; Look, Thomas; Begemann, Matthias; Zenke, Martin; Costa, Ivan G. (2019). "Identification of transcription factor binding sites using ATAC-seq". Genome Biology. 20 (1). doi:10.1186/s13059-019-1642-2. ISSN 1474-760X.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Mezger, Anja; Klemm, Sandy; Mann, Ishminder; Brower, Kara; Mir, Alain; Bostick, Magnolia; Farmer, Andrew; Fordyce, Polly; Linnarsson, Sten; Greenleaf, William (2018). "High-throughput chromatin accessibility profiling at single-cell resolution". Nature Communications. 9 (1). doi:10.1038/s41467-018-05887-x. ISSN 2041-1723.
- ^ Lareau, Caleb A.; Duarte, Fabiana M.; Chew, Jennifer G.; Kartha, Vinay K.; Burkett, Zach D.; Kohlway, Andrew S.; Pokholok, Dmitry; Aryee, Martin J.; Steemers, Frank J.; Lebofsky, Ronald; Buenrostro, Jason D. (2019). "Droplet-based combinatorial indexing for massive scale single-cell epigenomics". BioRxiv. doi:10.1101/612713.
- ^ Chen, Xi; Miragaia, Ricardo J.; Natarajan, Kedar Nath; Teichmann, Sarah A. (2018). "A rapid and robust method for single cell chromatin accessibility profiling". Nature Communications. 9 (1). doi:10.1038/s41467-018-07771-0. ISSN 2041-1723.
