ATAC-seq
用测序检测转座酶可及的染色质(ATAC-seq,英文全称为Assay for Transposase-Accessible Chromatin using sequencing)是用于检测基因组染色质可及性的分子生物学手段 [1]。此方法是MNase-seq、FAIRE-Seq和DNase-Seq的替代方法,于2013年发表[2][3][4]。对与表观基因组的分析,ATAC-seq比DNase-seq和MNase-seq都更快、更灵敏[2][3][4]。
描述
ATAC-seq 通过使用高活性突变型Tn5转座酶探测开放染色质来识别可访问的DNA区域,该转座酶可将测序接头插入基因组的开放区域[2][5]。天然存在的转座酶活性较低,ATAC-seq使用突变型转座酶具有高活性[6]。
在称为“标记化”的过程中,Tn5转座酶使用测序接头切割和标记双链DNA[7][8]。然后对标记的DNA片段进行纯化、PCR扩增,并使用下一代测序进行测序[8]。然后可以使用测序读取片段来推断可访问性增加的区域以及绘制转录因子结合位点和核小体位置的区域[2]。 在单核苷酸分辨率下,一个区域的读取片段个数与染色质的开放程度相关[2]。 ATAC-seq不需要像FAIRE-seq那样使用超声处理或苯酚-氯仿提取[9];不需要像ChIP-seq那样使用抗体[10];也不需要像MNase-seq或DNase-seq那样进行敏感的酶消化[11]。可以在三个小时内完成ATAC-seq的准备[12]。
应用
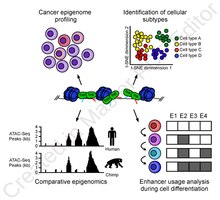
ATAC-Seq分析可以用于研究许多染色质可及性特征。最常见的用途是核小体定位实验[3],也可以可用于定位转录因子结合位点[13],适用于定位DNA甲基化位点[14],或与其他测序技术相结合[15] 。
高分辨率增强子映射的用途包括研究发育过程中增强子使用的进化差异(例如黑猩猩和人类之间的比较)[16]和揭示血细胞分化过程中使用的谱系特异性增强子图[17] 。
ATAC-Seq也被应用于定义人类癌症中全基因组染色质可及性情况[18],揭示了黄斑变性中染色质可及性的整体下降[19]。可以在 ATAC-seq 上运用计算足迹方法,以找到具有细胞特异性活性的细胞特异性结合位点和转录因子[13]。
单细胞 ATAC-seq
为了进行单细胞分析,有人已对ATAC-seq步骤进行了修改,以进行scATAC-seq(sc代表“单细胞”)。微流控技术可用于分离单个核并单独执行ATAC-seq反应[12]。通过这种方法,单个细胞在标记之前被微流体装置或液体沉积系统捕获[12][20]。一种不需要单细胞分离的替代技术是组合细胞索引。 [21]该技术使用条形码来测量数千个单个细胞中染色质的可及性;它可以在每个实验中生成10,000-100,000个细胞的表观基因组图谱[21]。但是组合细胞索引需要额外的、定制设计的设备或大量定制的、修改过的 Tn5[22]。最近,有人开发了称为sci-CAR的混合条形码方法,允许对单细胞的染色可及性和基因表达进行联合分析[23] 。
在进行scATAC-seq的计算分析时,可以以每个开放染色质区域的读取片段数建立计数矩阵。可通过伪多细胞的ATAC-seq数据的标准峰值来定义开放染色质区域。随后,可使用PCA进行数据降维,对细胞进行聚类[20]。scATAC-seq矩阵可能非常大(数十万个区域)并且非常稀疏,即不到3%的条目是非零的[24]。 因此,计数矩阵的填补是另一个关键步骤。与多细胞ATAC-seq一样,scATAC-seq可以找到调节因子,如控制细胞基因表达的转录因子。这可以通过查看围绕转录因子模式序列的读取片段个数[25]或足迹分析[24]来实现。
参考资料
- ^ Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature Methods. December 2013, 10 (12): 1213–8. PMC 3959825
 . PMID 24097267. doi:10.1038/nmeth.2688.
. PMID 24097267. doi:10.1038/nmeth.2688.
- ^ 2.0 2.1 2.2 2.3 2.4 Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Current Protocols in Molecular Biology. January 2015, 109: 21.29.1–21.29.9. PMC 4374986
 . PMID 25559105. doi:10.1002/0471142727.mb2129s109.
. PMID 25559105. doi:10.1002/0471142727.mb2129s109.
- ^ 3.0 3.1 3.2 Schep AN, Buenrostro JD, Denny SK, Schwartz K, Sherlock G, Greenleaf WJ. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Research. November 2015, 25 (11): 1757–70. PMC 4617971
 . PMID 26314830. doi:10.1101/gr.192294.115.
. PMID 26314830. doi:10.1101/gr.192294.115.
- ^ 4.0 4.1 Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harbor Protocols. February 2010, 2010 (2): pdb.prot5384. PMC 3627383
 . PMID 20150147. doi:10.1101/pdb.prot5384.
. PMID 20150147. doi:10.1101/pdb.prot5384.
- ^ Bajic M, Maher KA, Deal RB. Identification of Open Chromatin Regions in Plant Genomes Using ATAC-Seq. Plant Chromatin Dynamics. Methods in Molecular Biology 1675. 2018: 183–201. ISBN 978-1-4939-7317-0. ISSN 1064-3745. PMC 5693289
 . PMID 29052193. doi:10.1007/978-1-4939-7318-7_12.
. PMID 29052193. doi:10.1007/978-1-4939-7318-7_12.
- ^ Reznikoff WS. Transposon Tn5. Annual Review of Genetics. 2008, 42 (1): 269–86. PMID 18680433. doi:10.1146/annurev.genet.42.110807.091656.
- ^ Adey, Andrew. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biology. December 2010, 11 (12): R119. PMC 3046479
 . PMID 21143862. doi:10.1186/gb-2010-11-12-r119.
. PMID 21143862. doi:10.1186/gb-2010-11-12-r119.
- ^ 8.0 8.1 Picelli S, Björklund AK, Reinius B, Sagasser S, Winberg G, Sandberg R. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Research. December 2014, 24 (12): 2033–40. PMC 4248319
 . PMID 25079858. doi:10.1101/gr.177881.114.
. PMID 25079858. doi:10.1101/gr.177881.114.
- ^ Simon JM, Giresi PG, Davis IJ, Lieb JD. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nature Protocols. January 2012, 7 (2): 256–67. PMC 3784247
 . PMID 22262007. doi:10.1038/nprot.2011.444.
. PMID 22262007. doi:10.1038/nprot.2011.444.
- ^ Savic D, Partridge EC, Newberry KM, Smith SB, Meadows SK, Roberts BS, et al. CETCh-seq: CRISPR epitope tagging ChIP-seq of DNA-binding proteins. Genome Research. October 2015, 25 (10): 1581–9. PMC 4579343
 . PMID 26355004. doi:10.1101/gr.193540.115.
. PMID 26355004. doi:10.1101/gr.193540.115.
- ^ Hoeijmakers WA, Bártfai R. Characterization of the Nucleosome Landscape by Micrococcal Nuclease-Sequencing (MNase-seq). Chromatin Immunoprecipitation. Methods in Molecular Biology 1689. 2018: 83–101. ISBN 978-1-4939-7379-8. ISSN 1064-3745. PMID 29027167. doi:10.1007/978-1-4939-7380-4_8.
- ^ 12.0 12.1 12.2 Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. July 2015, 523 (7561): 486–90. Bibcode:2015Natur.523..486B. PMC 4685948
 . PMID 26083756. doi:10.1038/nature14590.
. PMID 26083756. doi:10.1038/nature14590.
- ^ 13.0 13.1 Li, Zhijian; Schulz, Marcel H.; Look, Thomas; Begemann, Matthias; Zenke, Martin; Costa, Ivan G. Identification of transcription factor binding sites using ATAC-seq. Genome Biology. 26 February 2019, 20 (1): 45. doi:10.1186/s13059-019-1642-2
 .
.
- ^ Spektor R, Tippens ND, Mimoso CA, Soloway PD. methyl-ATAC-seq measures DNA methylation at accessible chromatin. Genome Research. June 2019, 29 (6): 969–977. PMC 6581052
 . PMID 31160376. doi:10.1101/gr.245399.118.
. PMID 31160376. doi:10.1101/gr.245399.118.
- ^ Hendrickson DG, Soifer I, Wranik BJ, Botstein D, Scott McIsaac R, Simultaneous Profiling of DNA Accessibility and Gene Expression Dynamics with ATAC-Seq and RNA-Seq, Methods in Molecular Biology 1819, Springer New York: 317–333, 2018, ISBN 9781493986170, PMID 30421411, doi:10.1007/978-1-4939-8618-7_15
- ^ Prescott SL, Srinivasan R, Marchetto MC, Grishina I, Narvaiza I, Selleri L, et al. Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell. September 2015, 163 (1): 68–83. PMC 4848043
 . PMID 26365491. doi:10.1016/j.cell.2015.08.036.
. PMID 26365491. doi:10.1016/j.cell.2015.08.036.
- ^ Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. August 2014, 345 (6199): 943–9. PMC 4412442
 . PMID 25103404. doi:10.1126/science.1256271.
. PMID 25103404. doi:10.1126/science.1256271.
- ^ Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, et al. The chromatin accessibility landscape of primary human cancers. Science. October 2018, 362 (6413): eaav1898. Bibcode:2018Sci...362.1898C. PMC 6408149
 . PMID 30361341. doi:10.1126/science.aav1898.
. PMID 30361341. doi:10.1126/science.aav1898.
- ^ Wang J, Zibetti C, Shang P, Sripathi SR, Zhang P, Cano M, et al. ATAC-Seq analysis reveals a widespread decrease of chromatin accessibility in age-related macular degeneration. Nature Communications. April 2018, 9 (1): 1364. Bibcode:2018NatCo...9.1364W. PMC 5893535
 . PMID 29636475. doi:10.1038/s41467-018-03856-y.
. PMID 29636475. doi:10.1038/s41467-018-03856-y.
- ^ 20.0 20.1 Mezger A, Klemm S, Mann I, Brower K, Mir A, Bostick M, et al. High-throughput chromatin accessibility profiling at single-cell resolution. Nature Communications. September 2018, 9 (1): 3647. Bibcode:2018NatCo...9.3647M. PMC 6128862
 . PMID 30194434. doi:10.1038/s41467-018-05887-x.
. PMID 30194434. doi:10.1038/s41467-018-05887-x.
- ^ Lareau CA, Duarte FM, Chew JG, Kartha VK, Burkett ZD, Kohlway AS, Pokholok D, Aryee MJ, et al. Droplet-based combinatorial indexing for massive scale single-cell epigenomics. bioRxiv. 2019. doi:10.1101/612713
 .
.
- ^ Chen X, Miragaia RJ, Natarajan KN, Teichmann SA. A rapid and robust method for single cell chromatin accessibility profiling. Nature Communications. December 2018, 9 (1): 5345. Bibcode:2018NatCo...9.5345C. PMC 6297232
 . PMID 30559361. doi:10.1038/s41467-018-07771-0.
. PMID 30559361. doi:10.1038/s41467-018-07771-0.
- ^ Cao, Junyue; Cusanovich, Darren A.; Ramani, Vijay; Aghamirzaie, Delasa; Pliner, Hannah A.; Hill, Andrew J.; Daza, Riza M.; McFaline-Figueroa, Jose L.; Packer, Jonathan S.; Christiansen, Lena; Steemers, Frank J. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018-09-28, 361 (6409): 1380–1385. ISSN 0036-8075. PMID 30166440. doi:10.1126/science.aau0730
 (英语).
(英语).
- ^ 24.0 24.1 Li Z, Kuppe C, Cheng M, Menzel S, Zenke M, Kramann R, et al. scOpen: chromatin-accessibility estimation of single-cell ATAC data. bioRxiv. 2019-12-05: 865931. doi:10.1101/865931
 (英语).
(英语).
- ^ Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nature Methods. October 2017, 14 (10): 975–978. PMC 5623146
 . PMID 28825706. doi:10.1038/nmeth.4401.
. PMID 28825706. doi:10.1038/nmeth.4401.
